1. Background
Breast cancer is one of the most common malignancies leading to the death of women worldwide. Numerous factors, such as age, sex, obesity, estrogen levels, and family history are effective in increasing its prevalence (1). It is estimated that viruses are involved in about 16.1% of all human cancers (2). For example; HPV increases the risk of uterine cancer in cells that have decreased MTHFR activity (3) or human cytomegaloviruses (CMVs) are involved in the malignancy and progression of colorectal cancer (4). But no statistical relationship was observed between Epstein-Barr virus (EBV) and colorectal cancer (5). The results of cell line studies also showed that CMV infection could play a role in breast cancer (6). Overall, the high prevalence of viruses in breast cancer compared to normal samples indicates the role of viruses in the spread of breast cancer (7).
HCMV is an opportunistic double-stranded DNA virus that causes infections in the majority of the world's adult population. It can be transmitted through body fluids, such as saliva and breast milk, and like other herpesviruses, they remain hidden and permanent for part of their lifespan (8). These viruses exert a wide range of immunosuppressive effects. Virus-infected cells can prevent from being detected and eliminated by the immune system by orchestrating polarization of immunosuppressive type II macrophages, inhibiting antigen delivery, expressing T-cell inhibitory molecules, and possibly by inducing T-cell regulatory responses (Treg) (9). Human cytomegaloviruses exert their effects in vitro by deforming cells and deregulating pathways related to the pathogenesis of adenocarcinomas, especially cell cycle, mutagenesis, apoptosis, and angiogenesis (10).
HCMV is a beta-herpes virus with a high level of host specificity (11) that can target different types of cells in the body, including monocytes (12), macrophages, epithelial cells, endothelial cells, fibroblasts, stromal cells, nerve cells (stem and precursor), smooth muscle cells, and liver cells (11, 12), by which it can remain asymptomatic or cause a mild viral infection in the host (11). These opportunistic viruses cause severe and life-threatening diseases in people with a defective immune system, such as organ transplant recipients, cases with human immunodeficiency virus (HIV), and cancer patients (10).
2. Objectives
The aim of this study was to determine the level of CMV-IgM and CMV-IgG antibodies in the serum of individuals by examining the presence of viral genome in the breast tissue of women with breast cancer compared with healthy women.
3. Methods
Tissue and serum samples were prepared from those referring to 3 hospitals in Tehran, Iran (Shohadaye Tajrish, Imam Khomeini, and Khatam‑al Anbiya). Under the supervision of a specialist, 60 women with breast cancer and 60 healthy women (40 cases with fibroadenoma and 20 healthy samples) were selected from those who were subjected to surgery. Both groups were homogenized regarding age and did not have underlying diseases, such as cardiovascular disease and diabetes. Before sampling, questionnaires and informed consents were obtained from all individuals. Other pathological information of the patients was extracted from their medical records. The Ethics Committee of Shahid Beheshti University of Medical Sciences reviewed and approved the study protocol.
3.1. DNA Extraction & Real-time PCR
A part of the breast tissue subjected to surgery was placed in karyotype by a pathologist to be stored at -80°C, and after collecting all the samples, their DNA was extracted. The DNA of the samples was extracted by the salting-out method (13), and its quality and quantity were evaluated with agarose gel and Nano Drop. All extracted DNAs were then amplified with specific primers of the beta-globin gene that designed it (Forward: 5'-GAAGAGCCAAGGACAGGTAC-3' & Reverse: 5'-CAACTTCATCCACGTTCACC-3') and SYBR Green-based real-time PCR. The viral genome was also amplified using primers designed for part of the UL42 gene (Gene ID:3077440) (Forward: 5'-GTCCGAATGTTACGTCCAGC- 3'& Reverse:5'-ACTGCAGGGGTAAACGGTTA -3').
The amplification (100ng DNA, 10 μL SYBRTM Green 2X qPCR master mix & 1μL of each primer (10 pm/μL) & 7µL DDW) was performed with the ABI 7500 real-time PCR (Applied Biosystems, Life technologies) as follows: 95°C for 10 minutes, 35 cycles at 95°C for 15 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. Positive (Purified DNA was obtained Keyvan virology laboratory) and negative control samples (DNA-free samples) were used for the reactions. All reactions were performed in duplicate.
3.2. Serology
The serum of all subjects was taken 24 hours before surgery and kept at -80°C until testing. Using commercial kits (Immunolab GmbH, Germany), serum concentrations of IgM & IgG antibodies against HCMV in serum samples were determined to compare with real-time PCR results. The OD cutoff value was greater than 10 U/mL. According to the kit protocol, the results of the samples were measured by the ELISA technique in comparison with the kit standards.
3.3. Statically Analysis
Data analysis was performed using SPSS software version 23 and Graphpad prism version 9. The chi-square and Fisher’s exact tests were used for statistical comparisons between positive cytomegalovirus samples in cancer and control groups. The mean age, body max index (BMI), and serum levels of antibodies CMV IgG & IgM were also calculated in the case and control groups by using unpaired t-test and tables were drawn by Graphpad prism. In all analyses, results were reported as mean ± SD, and the P-value ≤ 0.05 level was considered significant.
4. Results
The mean age of patients with cancer was 54.70 ± 7.47 years (range 41 to 64 years) and the control subjects were 46.97 ± 6.36 years (range 41 to 62 years) (P =1.35 × 10-8). The mean BMI was 24.29 ± 1.85 Kg/m2 and 24.56 ± 2.76 Kg/m2 in cancer and control groups, respectively (P = 0.524). The demographic information of the subjects and also the number of positive samples for the viral genome are shown in Table 1.
| Variables | Case/Control | Number (%) | CMV Genome | P-Value | |
|---|---|---|---|---|---|
| Positive | Negative | ||||
| Age | 0.970 | ||||
| > 50 | Case | 42 | 11 | 31 | |
| Control | 20 | 2 | 18 | ||
| ≤ 50 | Case | 18 | 9 | 9 | |
| Control | 40 | 3 | 37 | ||
| BMI | 0.646 | ||||
| ≤ 25 | Case | 40 | 12 | 28 | |
| Control | 32 | 2 | 30 | ||
| > 25 | Case | 20 | 8 | 12 | |
| Control | 28 | 3 | 25 | ||
| ER | 0.311 | ||||
| Positive | Case | 43 | 16 | 27 | |
| Negative | Case | 17 | 4 | 13 | |
| PR | 0.989 | ||||
| Positive | Case | 36 | 12 | 24 | |
| Negative | Case | 24 | 8 | 16 | |
| Type of cancer | 0.234 | ||||
| ILC | 21 | 7 | 14 | ||
| IDC | 32 | 13 | 19 | ||
| DCIS | 4 | 0 | 4 | ||
| LCIS | 3 | 0 | 3 | ||
| Stage of cancer | 0.322 | ||||
| I | 6 | 3 | 3 | ||
| II | 14 | 3 | 11 | ||
| III | 20 | 5 | 15 | ||
| IV | 20 | 9 | 11 | ||
Correlation Between Distribution of CMV Genome and Clinic Pathological Indexes of Breast Cancer
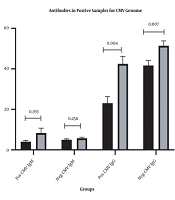
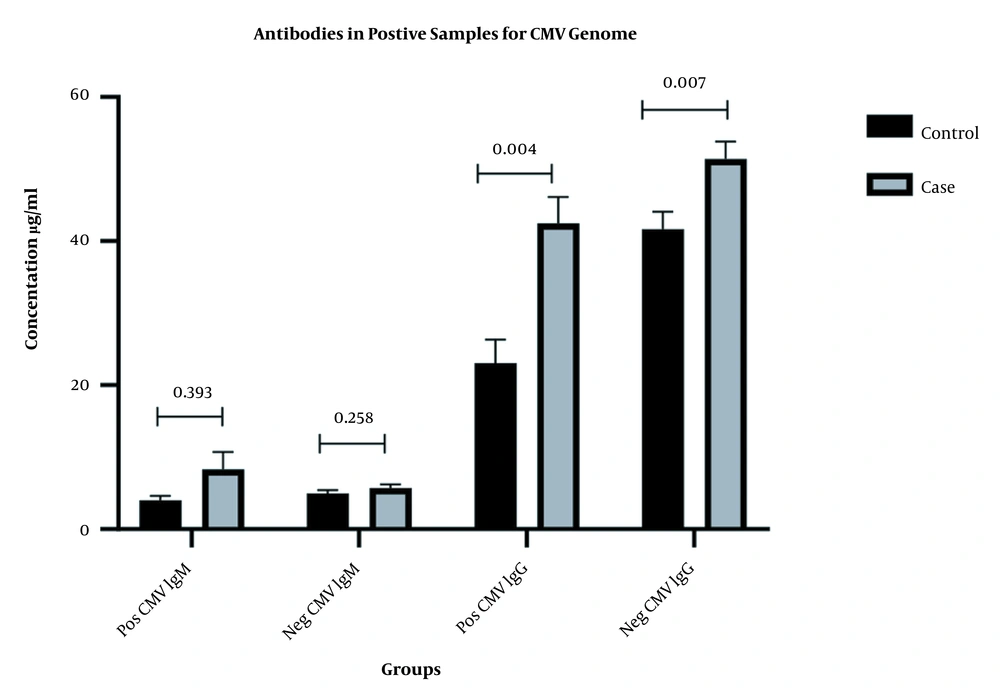
The viral genome was identified in samples of 20 patients with cancer and 5 control subjects (P = 0.001, OR: 5.50, CI95%: 1.90 - 15.89). CMV-IgM was observed in 5 patients with cancer and 3 controls (P = 0.464, OR:1.73, CI95%: 0.39 - 7.58). All cancer and control samples were positive for CMV-IgG antibody. However, the mean serum concentration of CMV-IgG was higher in the cancer group (48.27 ± 15.99 U/mL) than the control group (40.11 ± 18.01 U/mL) (P = 0.004). The mean serum concentration of the CMV-IgM antibody was higher in the cancer group (6.60 ± 6.75 U/mL) than the control group (4.92 ± 3.03 U/mL) (P = 0.099). Table 2 compares the serum levels of antibodies between samples with viral genomes and those without genomes in both groups (Figure 1).
| CMV IgM | CMV IgG | |||
|---|---|---|---|---|
| Pos CMV Genome | Neg CMV Genome | Pos CMV Genome | Neg CMV Genome | |
| Case | 8.32 ± 10.81 | 5.73 ± 3.11 | 42.48 ± 16.20 | 51.42 ± 15.16 |
| Control | 4.04 ± 1.30 | 4.99 ± 3.14 | 23.48 ± 16.20 | 41.66 ± 15.16 |
| P-value | 0.583 | 0.111 | 0.004 | 0.007 |
Mean Serum Concentration of Antibodies CMV IgM & CMV IgG in Positive and Negative Samples for Virus Genome
5. Discussion
Breast cancer is one of the most leading causes of cancer death in women. Viruses play an important role in developing breast cancer and other human cancers (11). The molecular results of 85 studies on the 3 viruses, including EBV, MMTV-LS, and HPV have shown that they play a role in breast cancer (14). In addition, the role of HCMV in developing or progressing breast cancer has been suggested (11). It has been reported that HCMV infection has been reported to modulate oncogenes-related signaling pathways (12).
IgM antibodies are useful to diagnose early infections with HCMV. However, these antibodies are also produced during viral reactivation or re-infection and possible cross-reactions with other viruses (15). In our samples, CMV-IgM antibodies were detected in the serum of 3 controls and 5 cancer patients. The mean concentration of antibodies was higher in the cancer group than in the control group, but this difference was not significant. However, the mean concentration of CMV-IgM antibody in the cancer group was higher than the control group, which this difference was statistically significant. In line with our results, Richardson also observed higher serum IgG levels in cancer samples than in control samples (16). Cox et al. also concluded that changes in the serum concentration of the CMV-IgG antibody could be used to diagnose the progression of breast cancer in some women. The increase in IgG antibody concentration may be due to new infection or viral reactivation, which is a risk factor for breast cancer (17).
Using a real-time PCR technique, of 60 cancerous breast tissues, 20 samples, and from 40 fibroadenoma samples, 5 samples were positive for HCMV genome. El Shazly et al. also identified the viral genome in 11 out of 60 cancer tissue samples and 1 out of 20 fibroblast samples. They also identified viral proteins in the samples. There was also a statistically significant difference in anti-CMV-IgG antibody levels between cancer and control samples (18). El-Shinawi et al. also found a statistically significant difference in HCMV-DNA in breast cancer tissues compared with non-cancerous tissues. In addition, the HCMV-IgG antibody titer was higher in cancer samples than in control samples (19). Harkins et al. detected the viral genome in 97% of neoplastic epithelial specimens and 63% of normal individuals through immunohistochemistry, in situ hybridization, PCR, and DNA sequencing techniques (20). Richardson et al. in a meta-analysis showed that 6 out of 7 studies reported breast cancer samples positive for CMV genome (7.4% to 100% of the samples). In addition, almost all studies performed to identify CMV using IHC and ISH techniques showed samples positive for the CMV genome (21). Mohamed et al. identified HCMV in metastatic breast cancer patients (1). Taher et al. also detected HCMV in 100% of cancer samples and 98% of sentinel lymph node (SLN) samples (22). In contrast, several studies found no association between HCMV infection and breast cancer (10, 23-25), or the CMV genome was identified only in 3% of healthy breast tissue samples using quantitative PCR (qPCR) (21).
The role of HCMV in cancer is unclear, but the interaction of viral proteins with cellular processes has been identified. IE proteins are regulatory proteins for the expression of viral and cellular genes that are synthesized during the virus life cycle. Regarding IE1 and IE2 proteins, several functions have been proposed, including cell deformation using the hit and run mechanism. In addition, other HCMV gene products may play a role in carcinogenesis by blocking cell differentiation, inducing chromosomal instability, DNA mutations, inducing migration, and angiogenesis (22). Given the potential role of HCMV in developing breast cancer, several factors may be associated with different results.
False-negative results, false-positive results, and the type of samples selected for comparison are the possible reasons for different results with PCR. Lacking quality evaluation of the studied DNA, Laser microdissection since the reduction in the number of viruses used, lack of hybridization due to detecting part of the gene or polymorphism in the viral genome, and the reduction of the virus during cell division causes the absence of viral DNA resulting in false-negative results. The false-positive results are more probable in the case of contaminating samples or using placental samples of cancerous and normal tissue (21). Other reasons include the limitations of molecular analysis, contamination with 2 or more viruses, and the use of the hit and run mechanism by viruses, which result in different results (23).
5.1. Conclusions
In the studied samples, a statistically significant relationship was observed between breast cancer and the presence of the virus genome compared with healthy tissues, and patients had higher levels of CMV-IgG antibody than controls. The absence of the viral genome in some cancer specimens may be due to the hit-and-run mechanism, by which the virus initiates cellular changes using a hit and decreases the viral genome while making sustained changes.