1. Background
Breast cancer is the most common cancer in women, accounting for 32% of all women's cancers. It is also the leading cause of cancer deaths in women after lung cancer and accounts for 19% of all cancer-related deaths in women (1).
The cause of breast cancer is unknown. However, epidemiological evidence has addressed the endocrine, environmental, and genetic factors (2).
Menopause can be considered one of the effective factors of breast cancer. It has been shown that early natural menopause and menopause by surgery can decrease the risk of breast cancer (3).
epidermal growth factor (EGF) and its tyrosine kinase receptor family play a role in the transmission of extracellular growth factor messages into the cell, regulating expression, and initiating proliferation. The EGF receptor (EGFR), known as ErbB1 or HER1, is a tyrosine kinase receptor and one of the first known receptors in this family. Three other members of the family have been also identified including ErbB2 (HER2), ErbB3 (HER3), and ErbB4 (HER4) (4).
HER2 plays an important role in cell growth and differentiation. The most important signaling pathway initiated by HER2 involves the establishment of MAPK and PI3K pathways. It is critical for cell survival as a key gene. HER2 gene amplification can lead to tumor malignancy (5).
Micro RNAs (miRNAs) are a family of small non-coding RNAs (21 – 25 nucleotides) that can inhibit mRNA translation. Micro RNAs can act as a biomarker for cancer and as a tumor suppressor or Oncogene through inhibiting the expression of cancer-related target genes. The expression of miRNAs varies in tumor and normal tissues. It also varies in different types of tumors (6, 7).
Many studies have shown the pattern of miRNAs expressed between healthy and diseased samples (8).
The expression of the miR-191 variable is associated with various diseases. Studies have shown that miR-191 is a promising new target for the diagnosis and treatment of cancer (9).
miR-22 has different effects on cancers ie, it not only acts as tumor suppressor miRNA but also as oncogenic miRNA and affects biological behaviors of cancer including proliferation, invasion, and metastasis (10).
Increased miR-22 can decrease Cyclin A2 protein, which affects cell division and protects over-proliferation (11).
2. Objectives
In general, utilizing a panel of miRNAs can increase sensitivity and specificity in the differential diagnosis of breast cancer in its early stages. Therefore, these markers have been investigated in the present study as early and differential markers in peripheral blood and tissues of patients with breast cancer.
3. Methods
3.1. Implementation Procedure
Blood and tissue samples were used in this study. The samples were collected in collaboration with the Cancer Institute of Masih Daneshvari Hospital, Shahid Beheshti University of Medical Sciences. Thus, 50 people with early stage I and II breast cancer were selected by a specialist. Also, 50 healthy individuals voluntarily participated in the study as a control group after completing the consent form. The patients and healthy individuals were considered in the same groups in terms of age and a range between 22 and 70 years old.
Also, 100 tissue samples were simultaneously collected from affected patients after obtaining written consent from them by a specialist and sent to the lab by a nitrogen tank. Among these tissue samples, 50 samples were obtained from the center of the tumor and 50 samples from the side tissues of tumors.
This study was approved by the Ethics Committee of Shahid Beheshti University of Medical Sciences (IR.SBMU.NRITLD.REC.1398.047).
3.2. Preparation of Blood and Tissue and RNA Extraction
After the selection of subjects, 2 mL of peripheral blood was taken through a blood syringe into a tube containing EDTA anticoagulant and immediately entered into the RNA extraction step.
The blood RNA extraction was carried out, using RNA Blood Mini Kit (Qiagen Cat no.52304) in accordance with Kit Protocol. The RNA concentration was measured, using NanoDrop technologies.
The tissue sample stored in a freezer at -80°C was placed on a plate. Then, 25 mg to 50 mg of fresh tissue was removed and 10 mg of it was cut, using a Bisturi scalpel or cutter Place two small pieces of tissue in a 2 mL microtube, and the tissue was pounded and homogenized to destroy the cells.
The tissue RNA was extracted through the standard method, using the RNA Extraction Kit of SINACLON Company in accordance with the manufacturer's instructions for tissue (solid) samples.
3.3. cDNA Synthesis and Conducting Real-Time RT-PCR
Vivantis Kit (Cat no.RTPL12) was used to make the cDNA. The 18srRNA reference gene was also used and a real-time PCR reaction was performed by Kit Sinaclone.
The specifications of primers used for real-time RT-PCR reaction have been presented in Table 1.
| Parameters | EGFR | 18s rRNA |
|---|---|---|
| Primer F | AAATCCTGCATGGCGCCGTG | TAACCCGTTGAACCCCATT |
| Primer length | 20 | 19 |
| Primer R | GGTGGTTCTGGAAGTCCATC | CCATCCAATCGGTAGTAGCG |
| Primer length | 20 | 20 |
PCR reaction was performed, using mixes, DNA, and primers with a final volume of 20.
PCR steps: 94°C for 10 minutes, 94°C for 20 seconds, 56°C for 40 seconds, 70°C for 40 seconds (35 cycles), and 70°C for 10 minutes.
For micro RNA, the ZIST ROYESH kit was also used for cDNA synthesis and real-time RT-PCR via Rotor-Gene – QIAGEN device. A housekeeping calibrator is needed to normalize the miRNA expression for each sample. In the present study, U6 was used as a housekeeping calibrator.
Temperature and time conditions are mentioned in the kit method. The results were evaluated based on melting and amplification curves.
In this study, the relative real-time PCR method was used to evaluate the difference in gene expression.
Mean expression levels of miRNAs and EGFR-mRNA were calculated, using the ΔΔCt method.
Parity-related expression was also determined, using the 2-ΔΔCt formula.
3.4. Statistical Analysis
SPSS 20 statistical software was used to analyze the results and calculate the variance, mean, and standard deviation. Paired t-test was used to analyze the differences between individuals of treatment and control groups in terms of the relationship between miRNA expression level and clinicopathological features. The significance level was considered at P ≤ 0.05.
4. Results
The understudy population included 50 healthy subjects and 50 subjects with breast cancer. These two groups were matched in terms of age variables. The groups were compared, using t-test in terms of mean age and no significant difference was observed in the mean age of the two groups. Therefore, it can be concluded that the age factor did not affect the two understudy groups (Table 2).
| Main Group | Age Range | Mean ± SD |
|---|---|---|
| subjects with breast cancer (50 individuals) | 25 - 65 | 47.32 ± 8.48 |
| Healthy subjects (50 individuals) | 25 - 64 | 45.78 ± 8.14 |
| P-value | 0.336 | |
All real-time RT-PCR reactions were performed so that 2 cDNA vials made from subjects with breast cancer and healthy subjects were tested to express the reference gene and understudy biomarkers. The results were interpreted based on the melt curve.
Analyzing the expression of understudy biomarkers:
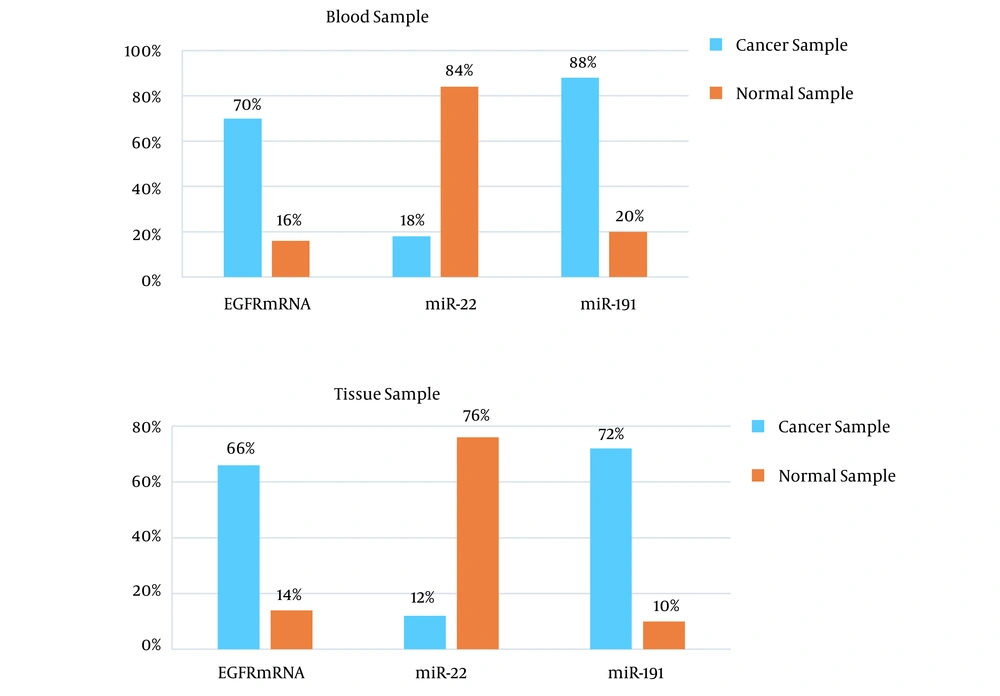
Peripheral blood EGFR mRNA biomarker was positive in 35 of 50 patients. The rate of this biomarker in the healthy group was 8 out of 50. Also, a positive EGFR mRNA marker was observed in tissue samples of the tumor center in 33 out of 50 patients. This rate was 7 out of 50 in the group of side healthy tissues. Using t-test, a significant difference was shown between the two groups (P < 0.001).
Biomarker miR-22 in peripheral blood was positive for 9 out of 50 patients and 42 out of 50 in healthy individuals. Also, the biomarker miR-22 in center cancer tissue was positive for 6 out of 50 in the patients' group. It was also positive in the side tissues for 38 out of 50 in the healthy group. Using t-test, a significant difference was shown between the two groups (P < 0.001).
Biomarker miR-191 in peripheral blood was positive for 44 out of 50 patients and 10 out of 50 in healthy individuals. Also, the biomarker miR-191 in center cancer tissue was positive for 36 out of 50 in the patients' group. It was also positive in the side tissues for 5 out of 50 in the healthy group. Using the t-test, a significant difference was shown between the two groups (P < 0.001) (Figure 1).
Calculating the difference in the expression level of biomarkers in two groups:
As was mentioned before, the Ct of each sample should be firstly determined. The relative difference of biomarkers expression between the samples of treatment and control groups was measured, using the ΔΔCt method (12-14).
In peripheral blood, the value of ΔΔCt for miR-191, miR-22, and EGFR mRNA biomarkers was -1.20, -0.55, and -1.44, respectively. In tissue cells, the value of ΔΔCt for miR-191, miR-22, and EGFR mRNA biomarkers was -1.11, -0.35, and -1.16, respectively.
On the other hand, the relative difference between the samples of treatment and control groups was obtained by the following formula:
Difference between expression of biomarkers = 2-ΔΔ Ct
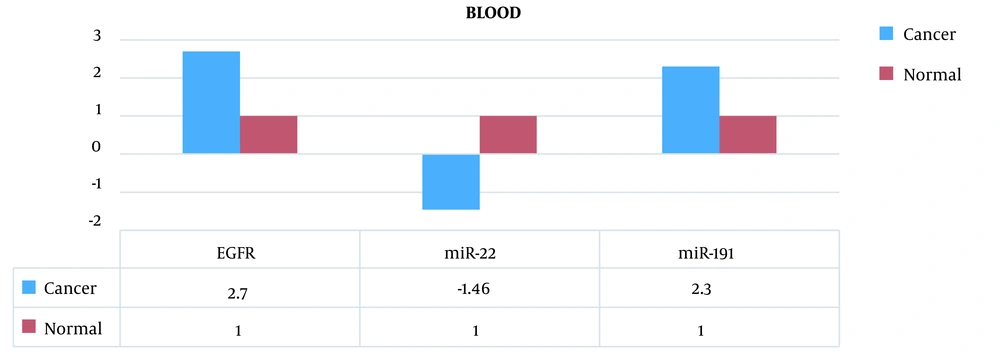
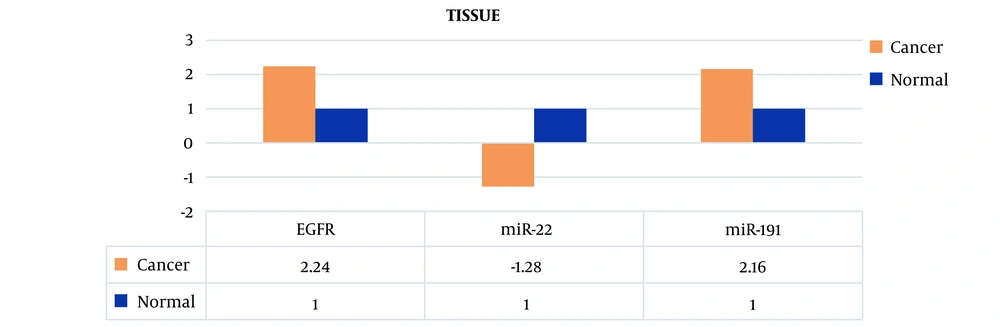
Therefore, the miR-191 expression level in peripheral blood of patients was 2.3 times higher compared to peripheral blood of healthy individuals. Also, the expression level of this biomarker in cancer tissue cells was 2.16 times higher compared to healthy tissue.
The miR-22 expression level in peripheral blood of patients was 1.46 times lower compared to peripheral blood of healthy individuals. Also, the expression level of this biomarker in cancer tissue cells was 1.28 times lower compared to healthy tissue.
The EGFR mRNA expression level in peripheral blood of patients was 2.70 times higher compared to peripheral blood of healthy individuals. Also, the expression level of this biomarker in cancer tissue cells was 2.24 times higher compared to healthy tissue (Figures 2 and 3).
5. Discussion
Breast cancer is the most commonly diagnosed cancer and the second leading cause of cancer-related deaths among women (15).
Various tumor markers are used to diagnose breast cancer including p21, HER2, ras, p53, integrin, CEA, and so on (16).
Studies have indicated the high expression of epidermal growth factors (EGFs) in breast cancer. Epidermal growth factor receptor (EGFR) has high expression in both cancer tissue and peripheral blood of cancer patients (17). In the present study, the expression of EGFR was also higher in peripheral blood and cancer tissue of cancerous samples compared to healthy samples.
Micro RNAs (miRNAs) are a family of small non-coding RNAs (21 – 25 nucleotides) that can inhibit mRNA translation. miRNAs modify gene expression after transcription through this mechanism (18).
The gathered evidence has shown that miRNAs can act as an Oncogene or as a tumor suppressor and measuring miRNA expression in malignancy can have diagnostic and prognostic consequences (19). However, there is still a lack of suitable molecular biomarkers for clinical applications in diseases.
The two markers of miR-22 and miR-191 have regulatory effects on cell proliferation, apoptosis, and migration through transcription factors.
Numerous studies have shown that MIR-191 and MIR-22 are suitable biomarkers for cancer diagnosis and prognostication. In the present study, the expression levels of MIR-191 and MIR-22 were investigated and their potential as a promising biomarker in breast cancer was discussed (20, 21).
In the present study, measuring the miR-191 expression levels in peripheral blood and tissues of cancerous and normal samples indicated an increase, so that the expression level of this biomarker in peripheral blood of patients was 2.3 times higher compared to healthy individuals and its expression level in the tissue of cancer cells was 2.16 higher compared to healthy tissue.
MiR-191 is highly conserved in several metazoan species and has been suggested as an important marker in Eukaryotes (22).
The diagnostic significance of Mir-191 in human serum has been demonstrated. The function of MiR-191 as an oncomiRNA has been shown in hepatocellular carcinoma and its increased expression enhances cell growth and decreases apoptosis; this result is consistent with the results of the present study (20). High expression of MIR-191 is also significantly associated in adult AML patients (23). In the study of Nagpal et al. (24), mir-191 is responsible for promoting tumor markers in ER+ of breast cancer cells. On the other hand, Di Leva et al. (25) showed that mir-191/425, can reduce proliferation, tumorigenesis, and metastasis in ER- (estrogen receptor -) of breast cancer cells. MiR-191 is an ER regulatory miRNA and acts as an important estrogenic mediator. miR-191 regulates the progression of cell proliferation in breast tumors by crossing from G1 / S phase to G2 / M phase. In the present study, the level of mir-191 expression was also increased in both tissue and peripheral blood.
It has been also reported that miR-191 acts as an oncogene in breast cancer and exhibits increased expression and induces cell proliferation (24). It has been also emphasized in the present study that miR-191 as a new biomarker can represent a potential prognosis in breast cancer.
It has been shown that mir-22 can lead to reduced cancer growth and invasion and impair tumor progression by inhibiting the cell cycle in a variety of ways. The matter indicates that mir-22 can act as a tumor suppressor in response to various carcinogens to reduce or inhibit cancer incidence and progression (10). This is in accordance with the results of the present study. The study conducted by Wang et al. also indicated a decrease in mir-22 expression level so that mir-22 inhibits tumor proliferation, invasion, and metastasis (10).
In the present study, measuring the miR-22 expression levels in peripheral blood and tissues of cancerous and normal samples indicated a decrease, so that the expression level of this biomarker in peripheral blood of patients was 1.46 times lower compared to healthy individuals and its expression level in the tissue of cancer cells was 1.28 lower compared to healthy tissue.
In a study, it has been shown that MIR-22 expression inhibits proliferation, cell growth, and migration, and hence, the decreased expression of MIR-22 leads to proliferation and progression to cancer (21). The decreased expression of this biomarker in blood peripheral and tissue has been also shown in the present study.
5.1. Conclusions
In summary, overexpression of miR-191 and EGFR-mRNA and decreased expression of miR-22 can play an important role in the prognostication of patients with cancer. The results of the present study showed that miR-22 and miR-191 can be important molecular markers for the prognosis of breast cancer. Of course, there were some limitations in this study, such as access to clinical samples and preparation of appropriate laboratory materials, and therefore more clinical studies are needed to confirm these findings.