1. Background
Primary bone tumors are considered a leading cause of death and impose a burden of morbidity on patients, worldwide. Among the different types of primary bone tumors, osteosarcoma is a common type with the ability to induce a high degree of malignancy, metastasis at early stages, poor prognosis, and high rate of progression in individuals with different age groups (1). Current therapeutic and detection approaches failed to improve the survival rate of osteosarcoma and its considerable death rate remains (2). Notably, the combined therapy approaches such as surgery in combination with multiple chemotherapy regimens reached a remarkable improvement in the patient’s survival rate; however; chemoresistance is observed in some patients and worth to be considering (3). Determining how cell death pathways can overcome the problem of resistance to chemotherapy in these patients can make treatment more effective. It has recently been shown that other pathways of cell death, including autophagy and necroptosis, can be as effective in the induction of death by treatment regimens as apoptosis (3). As the increase in the rate of cell proliferation compared to cell apoptosis results in neoplasm and excess cell growth, recent efforts concentrated on targeting cell death or proliferation pathways to develop more efficient anti-cancer therapies (4, 5). Among the cell death pathways, autophagy critically affects cell proliferation and differentiation that can act as a tumor suppressor and influence cell fate (6). During autophagy as a catabolic process, the formation of a double membrane structure, namely autophagosomes is an initial and critical step and a significant number of proteins are involved in proceeding the pathway (7). Among the autophagy mediators, the human autophagy-associated (ATG) proteins are involved in autophagosome formation and consist of several proteins including ATG12, ATG8, ATG7, and ATG5 that ultimately, ATG5, microtubule-associated protein 1A/1B-light chain 3 (LC3) and Beclin 1 (BECN1) account as main mediators to engulf cytoplasmic components (8). Besides, necroptosis, as a cell death pathway that acts regardless of caspase activation, is dominant during apoptosis-lacking conditions. The swelled cell with impaired mitochondrial function and permeabilized plasma membrane and leaking of cytoplasmic content to the extracellular space are the main phenotypic features of necroptosis (9, 10). Receptor-interacting protein kinases (RIPKs) are serine/threonine kinases that regulate the necroptosis pathway (11). Briefly, phosphorylation of mixed lineage kinase domain-like (MLKL) by receptor-interacting protein kinase-3 (RIPK3) induces the formation of RIPK1–RIPK3-MLKL complex, also called the ‘necrosome’, mediates accumulation of reactive oxygen species (ROS), permeabilization of the cell membrane and cellular adenosine triphosphate (ATP) depletion (9). Multiple lines of pieces of evidence support the fact that deficient cell death serves as the main event in early cancer cell development and growth also responsible for response to therapy and chemotherapy resistance (12). Targeting apoptotic pathway markers has been shown to inhibit the growth of osteosarcoma cancer cells, but the problem of resistance to treatment in this disease has not yet been resolved (3). As shown in previous studies, among the inducers of autophagy and necroptosis pathway, ATG5, BECN1 and LC3 (for autophagy) (13) and RIPK1, RIPK3 and MLKL (for necroptosis) (14, 15) are the most important biomarkers of these pathways that play a central role in pathway induction, and are usually selected in studies to examine these pathways.
2. Objectives
Regarding the potential role of different cell death pathways in cancer cell fate and their potent capacity to be triggered as anti-cancer agents, the current study aimed at evaluating the expression pattern of ATG5, BECN1, LC3, as main mediators of autophagy and RIPK1, RIPK3 and MLKL, as main mediators of necroptosis in osteosarcoma tumors and healthy bone tissues to provide evidence regarding the status of these pathways in bone cancer.
3. Methods
3.1. Patients and Sample Collection
A total of 60 bone tumor tissues including 40 malignant tumors and 20 benign tumors as well as 20 tumor margins were collected and enrolled in the current study with local ethical approval and informed consent (noted in the ethics statement). The sample size was calculated, using statistical consulting and based on the results of previous studies (16). The tumor and tumor margin samples were resected from patients who were subjected to surgery at the Shafa Orthopedic Hospital. The tumor and normal bone tissue collection and processing were followed as in our previous study (17). Briefly, following tumor resections, tissues were divided into 2 sections; one for histological assessments by the pathologist and the rest for gene analysis, which was stored immediately at -80°C until usage. The tumor margins were taken as control tissues and were verified by an expert pathologist. The clinic-pathological features of the patients with bone tumors are summarized in Table 1.
| Variables | Benign (n = 20) | Malignant (n = 40) |
|---|---|---|
| Age (y) | ||
| > 20 | 4 (20) | 17 (42.5) |
| 20 - 40 | 8 (40) | 14 (35) |
| 40 - 60 | 6 (30) | 8 (20) |
| < 60 | 2 (1) | 1 (2.5) |
| Sex | ||
| Female | 12 (60) | 26 (65) |
| Male | 8 (40) | 14 (35) |
| Chemotherapy status | ||
| Chemotherapy + | 28 (70) | |
| Chemotherapy - | 12 (30) | |
| Grade | ||
| Low grade (grade I and II) | 25 (62.5) | |
| High grade (grade III and IV) | 15 (37.5) |
a Values are expressed as No. (%).
3.2. RNA Extraction
To evaluate the gene expression pattern, RNA extraction was carried out based on the method recommended by the TRIzol-reagent manufacturer protocol. Briefly, the manual homogenizer was used to disrupt the tissue cells. The chloroform and isopropanol were applied subsequently following several incubations and centrifugations in cold conditions to isolate the RNA content. The extracted RNA was precipitated, using 75% ethanol (EtOH), and stored at -80°C following adequate washing steps. To evaluate the quantity of the extracted RNA, the absorbance of the extraction was assessed by a nanodrop spectrophotometer (NanoDrop Technologies). Briefly, 1 - 2 microliters of extracted RNA were applied and the absorption values at 260 nm (represents the presence of nucleic acids), 280 (represents the presence of proteins), and 230 (related to organic solvents) were assessed. The absorption ratios at 260/280 determine the amount of protein contamination: a ratio of > 1.8 indicates appropriate purity of RNA and the absorbance ratios at 260/230 determine the degree of contamination with organic solvents: a ratio of > 2 indicates no contamination with such solvent. The final concentration of RNA was reported as micrograms per microliter. Subsequently, to indicate the RNA integrity and purity, RNA was electrophoresed, using 1% agarose gel. RNA has a negative charge due to the presence of phosphate groups and moves to the anode in the presence of an electric field. The speed of this movement depends on the mass of the molecule relative to the charge. If the mRNA molecules are of the desired integrity, only 2 bands corresponding to the 18S and 28S RNAs will be observed on the gel electrophoresis.
3.3. cDNA Synthesis and Real-time PCR
The amount of 1 μg of the extracted RNA was applied for cDNA synthesis, using PrimeScript First Strand cDNA Synthesis Kit based on the manufacturer’s instructions. Briefly, RNA (1000 ng), 1 μL of dNTPs (10 mM), 0.5 μL reverse transcriptase, and either 1 μL of oligo (dT) primer, 1 μL of the random hexamer, or a combination of both were combined and brought to a final volume of 10 μL with RNase-free water. Samples were incubated at 42°C for 25 minutes and 85°C for 7 seconds and chilled on ice. The cDNA of each sample was used for real-time PCR reaction to quantitatively determine the level of mRNA expression of ATG5, BECN1, LC3, RIPK1, RIPK3, and MLKL genes, using Sybr Green High ROX Mastermix. The specific primers were designed for each gene and the sequences of the primers. The size of amplification products is illustrated in Table 2. The β-actin (ACTB) was used as an endogenous control (housekeeping gene) to normalize differences in the amount of total RNA in each sample. The running program was as 14 minutes at 95°C, followed by 40 cycles of 15 seconds at 95°C, 20 seconds at 55°C, and 35 seconds at 60°C. Real-time PCR was performed, using ABI Prism 7000 Sequence Detection System (Applied Biosystems, USA) and all samples were analyzed in duplicate. The comparative CT (2-ΔCt) method was applied for gene expression analysis.
| Symbol | Gene Name HGNC | Product Length | Forward Primer (5’ → 3’) | Reverse Primer (5’ → 3’) |
|---|---|---|---|---|
| ACTB | BRWS1, PS1TP5BP1 | 137 | GATCTCCTTCTGCATCCTGT | TGGGCATCCACGAAACTAC |
| RIPK1 | IMD57, RIP, RIP-1, RIP1 | 178 | GTGCTGAAAGCCGAGATGAGTA | GCTTGTTTTGAGCTGTAGCCTG |
| RIPK3 | RIP3 | 167 | ATCCAGTAACAGGGCGACCG | TTGCGAACCTACTGGTGGGG |
| MLKL | Hmlkl | 109 | CTTTGAGGCAGTATTTACCACC | TCCCTTAGCAGAATCCACG |
| LC3A | MAP1LC3A | 135 | AGTTGGTCAAGATCATCCG | CGTCCTCGTCTTTCTCC |
| BECN1 | BECN1 | 131 | GACACGAGTTTCAAGATCCTG | TCAATAAATGGCTCCTCTCCTG |
| ATG5 | ATG5 | 176 | TGGAGTAGGTTTGGCTTTGG | AATAGTATGGTTCTGCTTCCCT |
Abbreviations: ACTB, β-actin; RIPK, receptor-interacting protein kinases; MLKL, mixed lineage kinase domain-like; LC3, microtubule-associated protein light chain 3; BECN1, Beclin 1; ATG5, autophagy-associated protein 5.
3.4. Statistical Analysis
The gene expression analysis was calculated, using the CT (2-ΔCt) method, and the differences in each gene expression between groups were statistically determined, using a t test and one-way analysis of variance (ANOVA). The normal distribution of data in each analyzed group was assessed, using Kolmogorov-Smirnov analysis. The correlation of gene expression levels with the patient’s clinic pathological features was assessed, using the Pearson correlation coefficient test. Graph Pad Prism version 8 (Graph Pad Software, San Diego California) was used for the calculation of all statistics. P values < 0.05 (two-sided) were considered statistically significant.
4. Results
4.1. The Expression Pattern of Autophagy Main Mediators in Osteosarcoma Bone Tumors
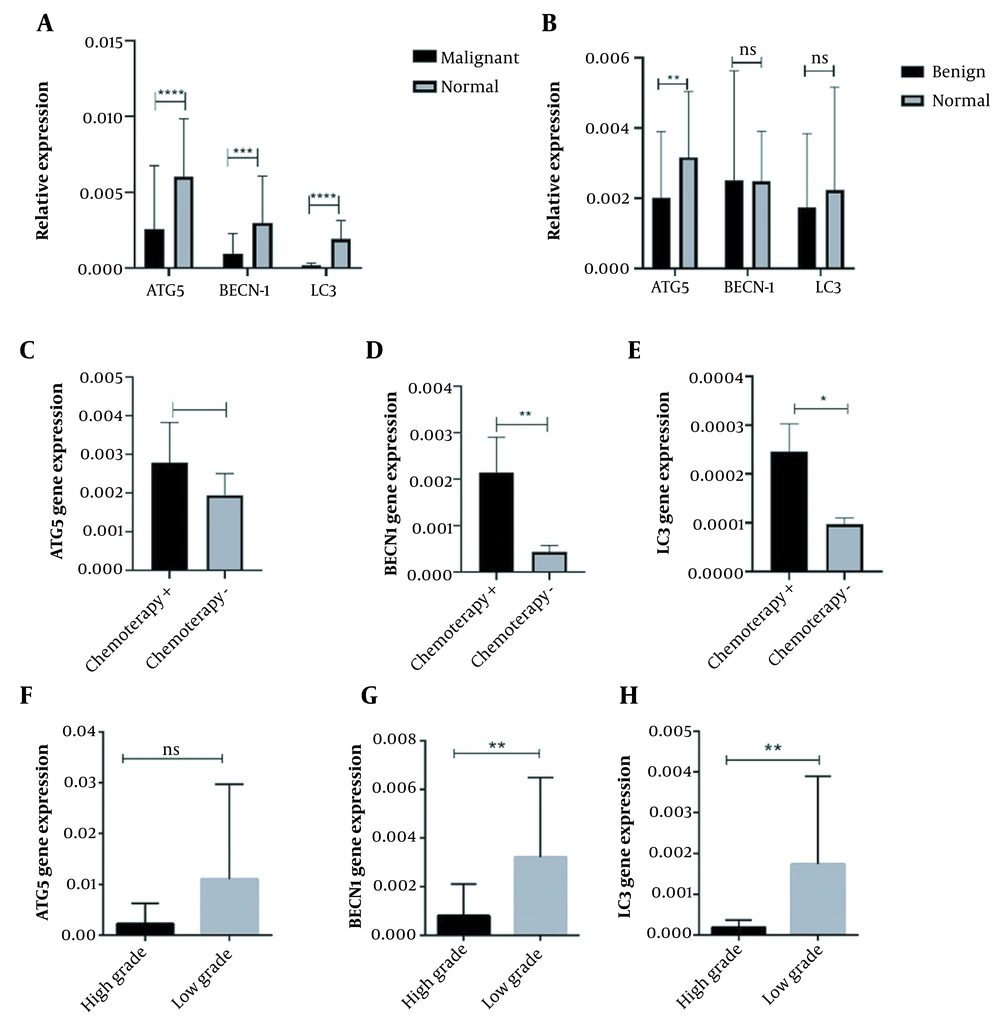
As is illustrated in Table 1 regarding the demographic features of our patients, 40% of patients with benign bone tumors were between 20 to 40 years of age while malignant bone tumors occurred mostly below the age of 17, and the mean and standard deviation of the patient’s age was 29.06 ± 14.72. Also, 60% and 57% of the patients were female in benign and malignant bone tumors, respectively. Additionally, 62.5% of malignant tumors were in grades I and II, while 37.5% of malignant bone tumors were in grades III and IV of malignancy. To clarify whether a chemotherapy regimen can trigger autophagy and necroptosis cell death pathways in osteosarcoma patients, both types of patients with and without a history of chemotherapy were enrolled in this study. Among patients with malignant bone tumors, 70% of patients received chemotherapy treatment before the surgery, while 30% of patients did not receive a chemotherapy regimen. Notably, the standard combination of doxorubicin, cisplatin, and methotrexate was applied to osteosarcoma patients as chemotherapy approaches. To better show and compare the results, the expression of genes in the group of malignant and benign tumors and healthy bone tissue were examined and compared. Also, considering that tumor grade and history of chemotherapy are two important and decisive features in the clinical condition of patients with malignant tumors, we separately compare the expression of each gene in malignant tumors with or without chemotherapy. Also, the expression pattern of each gene in malignant tumors with high or low grades was compared and shown in the graphs. Regarding the expression pattern of autophagy mediators, our data showed that the expression pattern of ATG5, BECN1, and LC3, as main autophagy mediators significantly decreased in malignant osteosarcoma tumors compared to tumor margin tissues (P < 0.0001, P = 0.0037, and P < 0.0001, respectively) (Figure 1A). Accordingly, the reduced expression level was more prominent for ATG5 and LC3 in osteosarcoma tumors. The attenuated level of ATG5, BECN1, and LC3 gene expression was also observed in benign osteosarcoma tumors compared to tumor margin tissues, while the observed differences were not statistically significant (Figure 1B). Our data revealed that the mRNA expression level of ATG5, BECN1, and LC3 was enhanced in tumors following the chemotherapy regimen, which was significant for BECN1 (P = 0.0037) and LC3 (P = 0.0105) when compared to tumors of patients, who were not under chemotherapy regimen (shown in Figure 1C - E). Notably, tumor grading is applied in a process of diagnosis to define the invasive behavior of the tumors and high-grade tumors (grades 3 and 4) are characterized as those tumors with moderate or poorly differentiated cells compared to normal bone cells. In the current study comparing the high-grade tumors (3 or 4) to low-grade tumors (1 or 2), it was revealed that the expression level of ATG5, BECN1, and LC3 was considerably reduced in high-grade osteosarcoma tumors compared to low-grade tumors (Figure 1F - H). The difference in BECN1 and LC3 expression between high-grade tumors and low-grade tumors was statistically significant (P = 0.0063 and P = 0.0078, respectively), while the difference in ATG expression between the mentioned groups was nearly significant (P = 0.055).
The expression pattern of autophagy mediators in osteosarcoma tumor tissues. The autophagy-associated protein 5 (ATG5), Beclin 1 (BECN1), and microtubule-associated protein light chain 3 (LC3) expression levels were evaluated in osteosarcoma bone tumor tissues, and the decreased level of ATG5, BECN1, and LC3 expression was observed in A, malignant tumors versus normal bone tissues also B, benign bone tumors versus tumor margins except for BECN1. The C, ATG5; D, BECN1; and E, LC3 expressions were increased in chemotherapy-received tumors compared to tumors without a history of chemotherapy. The expression level of F, ATG5; G, BECN1; and H, LC3 were reduced in high-grade tumors. The statistical differences between groups are shown as asterisk (* P < 0.05, ** P < 0.01, **** P < 0.0001).
4.2. The Expression Pattern of Necroptosis Main Mediators in Osteosarcoma Bone Tumors
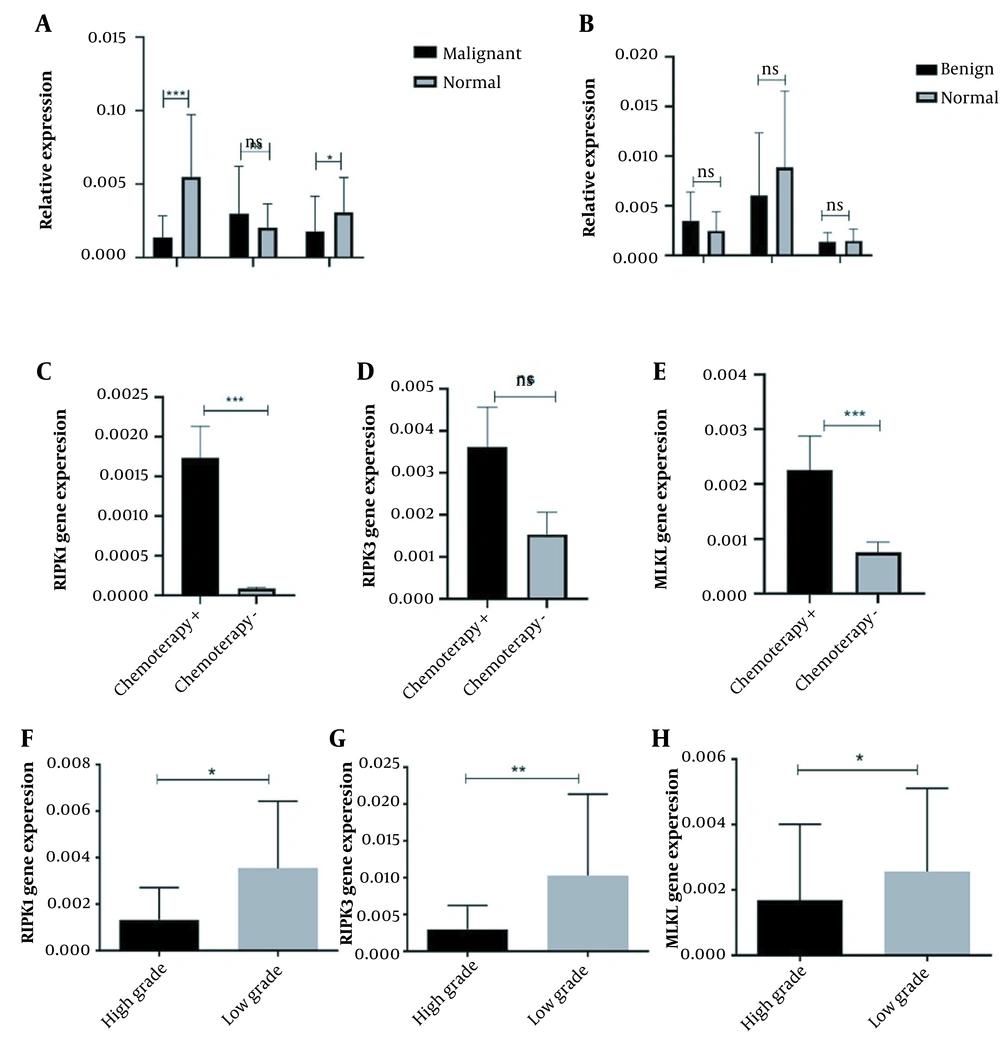
To evaluate the possible role of the necroptosis pathway as an alternative programmed cell death pathway in cells, the mRNA level of RIPK1, RIPK3, and MLKL was assessed in osteosarcoma tumors. Based on our data, it was shown that the expression level of RIPK1 (P = 0.0028) and MLKL (P = 0.0025) was significantly decreased in malignant osteosarcoma tumors compared to tumor margins (Figure 2A), while the increased expression of RIPK3 was not remarkable in malignant tumors compared to tumor margins. Notably, in benign bone tumors, the expression level of RIPK1 and MLKL was decreased in tumors compared to tumor margins, which were considered for MLKL than RIPK1 and RIPK3; however, the observed differences were not statistically significant (Figure 2B). Interestingly, the expression level of RIPK3 increased in benign tumors compared to tumor margins, while the difference was not significant (Figure 2B). Considering the effect of the chemotherapy regimen on necroptosis mediators, our data revealed the higher expression of RIPK1, RIPK3, and MLKL in tumors of patients, who received the chemotherapy regimens compared to the tumors without a history of chemotherapy (Figure 2C, D and F). In accordance, the higher expression level of RIPK1 (P = 0.0002) and MLKL (P = 0.0008) was statistically significant in tumors of patients, who received the chemotherapy regimens compared to tumors of patients without receiving chemotherapy regimen, while the difference was not considered for RIPK3. Based on our data, the expression level of RIPK1 (P = 0.0107), RIPK3 (P = 0.0073), and MLKL (P = 0.0423) was significantly decreased in high-grade osteosarcoma tumors compared to low-grade tumors, indicating the fact that suppression of necroptosis pathway might facilitate the proliferation of osteosarcoma tumor cells (Figure 2F - H).
The expression pattern of necroptosis mediators in osteosarcoma tumor tissues. The receptor-interacting protein kinases (RIPK) 1, RIPK3, and mixed lineage kinase domain-like (MLKL) expression levels were evaluated in osteosarcoma bone tumor tissues. A, The expression level of RIPK1 and MLKL was decreased, while RIPK3 showed a non-significant increase in malignant tumors versus normal bone tissues; B, The expression level of RIPK1, RIPK3, and MLKL did not significantly change between benign bone tumors and tumor margins. The expression level of C, RIPK1; D, RIPK3; and E, MLKL was increased in chemotherapy-received tumors compared to tumors without a history of chemotherapy. The expression level of F, RIPK1; G, RIPK3; and H, MLKL were decreased in high-grade tumors. The statistical differences between groups are shown as an asterisk (* P < 0.05, ** P < 0.01, *** P < 0.001).
5. Discussion
The frequent mutations in apoptotic machinery in many types of cancers account for important causes of chemoresistance (13, 18). Therefore, targeting other programmed cell death pathways in chemotherapeutic approaches may open up more efficient and promising anti-cancer therapeutic strategies (19). Among cell death pathways, autophagy may display dual behavior regarding cell fate, it can contribute to cell death or cell survival depending on the cell condition, which results in an increase or decrease in the sensitivity of the tumor to chemotherapy regimen (20). It was revealed that defects of the autophagy pathway in tumors caused a decrease in the immune system response of the tumor to chemotherapy agents (19). In accordance, the problem of tumor recurrence and chemoresistance in osteosarcoma, as a malignant primary bone tumor, provoked us to provide piece of evidence regarding the expression pattern of autophagy main mediators in osteosarcoma tumors. Based on our data, the ATG5, BECN1, and LC3 mRNA levels were decreased in osteosarcoma tumor tissues compared to tumor margin tissues. The expression pattern of ATG5, BECN1 and LC3 genes revealed a positive correlation with tumor malignancy since the expression level of ATG5, BECN1 and LC3 showed a significantly lower level compared to benign tumors. In support of our data, it was revealed that BECN1 expression was decreased in osteosarcoma tumors compared to normal bone tissues; however, treatment of MG63 cells with a high dose of cisplatin as a chemotherapy agent resulted in increases in the expression level of BECN1, which indicates the possible involvement of autophagy in osteosarcoma pathogenesis (21). In addition, it was revealed that the ratio of LC3-II/LC3-I in the osteosarcoma cell line was lower compared to normal osteoblast, indicating a decrease in the activity of the autophagy pathway in the bone tumor, which was accompanied by up-regulation of phosphoinositide 3-kinases (PI3K) in the tumor cells (22). In line with our data, it was shown that triggering autophagy, using herbal-based compounds, promotes the chemo-sensitivity of osteosarcoma cells (3). On the other hand, defects in necroptosis as a programmed cell death pathway are related to inflammation and cancer manifestations and can account for an appealing target for anti-cancer strategies (23). The status of the necroptosis pathway in osteosarcoma tumors was vague and data of the current study revealed that the expression levels of RIPK1 and MLKL were decreased significantly in malignant tumors compared to tumor margin tissues; also the RIPK3 and MLKL down-regulation was observed in benign tumors compared to tumor margin tissues, while RIPK1 expression showed a slight increase in Benign tumors versus normal tissues. However, a significant change in expression pattern was not observed for RIPK1, RIPK3, and MLKL in benign tumors. So far, few studies have been presented on the role of the necroptosis pathway in the pathogenesis of osteosarcoma (3). Among them, it was shown that shikonin, Chinese medicinal herbs, and a well-documented necroptosis inducer (15) can trigger osteosarcoma cell death in an inducing RIP1 and RIP3 dependent manner (24). In accordance, it was shown that the transcription of RIPK3 is regulated by genomic methylation since in the presence of hypo-methylation agents, the attenuated expression of RIPK3 was restored and the sensitivity to chemotherapy treatments was increased (25). Therefore, the methylation level of RIPK3 can affect its mRNA level in our study that should be verified by future mechanistic investigations. Also, based on multiple evidence, variations observed in the expression pattern of RIPK3 might be due to the tumor type, tumor stage, and tumor features (26, 27). Given the important role of the necroptosis pathway in cancer cell growth, the association of MLKL reduced expression with the patient’s poor survival was observed in pancreatic adenocarcinoma (9). The major obstacles to chemotherapy regimens are an aberrant expression of programmed cell death inhibitors or extreme low expression of pro-apoptotic mediators in tumors, which may cause apoptosis evasion and chemoresistance (28). Accordingly, focusing on other alternatives to programmed cell death may provide better solutions for more effective treatments (8). Since improvement in chemoresistance problems may improve survival rate in osteosarcoma patients (3), the correlation of chemotherapy regimen on autophagy and necroptosis mediators’ expression patterns was surveyed in the current study. Based on our data, the patients who received chemotherapy regimens, showed higher expression levels of BECN1, LC3, RIPK1, and MLKL compared to the rest of the patients. In support of our data, it was revealed that cisplatin can stimulate death pathways including autophagy and necroptosis, which may vindicate our data.
5.1. Conclusions
Taken together, our data provide pieces of evidence regarding the reduced expression of autophagy and necroptosis as main mediators in osteosarcoma tumors, which were correlated with tumor malignancy. Also, the increased expression level of autophagy and necroptosis main mediators in patients under chemotherapy regimen indicates that these pathways can be effective in responding to chemotherapy and can be considered for further investigations. Therefore, in situations where patients do not respond to conventional chemotherapeutic regimens, necroptosis and autophagy pathways can be targeted to remove the resistance barrier and eliminate cancer cells more effectively in osteosarcoma.