1. Background
Gastric cancer (GC), the second most common malignancy globally and with 3% to 10% of all cancer-related mortality, has long been a life-threatening disease sophisticated by limitless factors interfering with its treatment (1-3). Despite the valuable efforts for improving the treatment strategies for GC cancer, still, a considerable proportion of patients lose their lives mainly due to the induction of drug resistance (4, 5). Even the addition of trastuzumab to the first-line treatment of cancer, either as a single agent or in combination with paclitaxel, was not successful enough to prolong the overall survival of the patients (6). However, the recent advances in the novel strategies such as gene therapy and its success in the treatment of a wide range of inherited and acquired human diseases have shed a ray of hope for the treatment of cancer patients, especially GC patients (7).
Introducing genes by plasmids into the genome of tumoral cells and generating appropriate quantities of the gene product to induce compulsory cell death mediated by overexpression of a specific tumor-suppressor gene is an appealing application of gene therapy in the field of cancer therapy (8). Among a wide variety of genes that could be fused with the genome, death-associated protein kinase 3 (DAPK3) is one of the most investigated ones (9, 10). Through interplaying with a variety of signaling pathways, DAPK3, the third member of the DAPK enzymes family, could regulate a vast array of cellular functions, including caspase-dependent apoptotic cell death, autophagy, cell adhesion, as well as cell migration (11). The biological role of DAPK3 in cells has been gradually investigated in recent years (12). Death-associated protein kinase 3 is pro-apoptotic and carries out this function eithervia type I (caspase-dependent) apoptotic or type II (caspase-independent) autophagic cell deaths (12). Death-associated protein kinase 3 also plays an important role in mediating inflammatory signals such as L13a (ribosome protein), ERK, and inhibiting interferon-activated translation (IFN) (12). The contributory role of DAPK3 has been examined in different cancer cell types; however, in many cases, there are conflicting results. Although numerous investigations are emphasizing the anti-tumor activity of DAPK3, other studies declare that the overexpression of DAPK3 in cancer cells is in favor of tumor cell survival (13). Despite several studies, the exact molecular activity of DAPK3 and the signals that regulate DAPK3 activity in cancer cells have not been fully elucidated and more research needs to be conducted (12). Nevertheless, the data regarding the effect of this protein on cancer cells through the apoptotic and autophagy pathways are preliminary (12). Therefore, in the present study, we investigated the effect of DAPK3 overexpression, using the non-viral PEGFPN1 vector on a well-known human gastric adenocarcinoma cell line and examined its overexpression impacts on gastric carcinoma.
2. Methods
This study protocol was confirmed by the Ethics Committee of Shahid Beheshti University of Medical Sciences (48-97/1/26).
2.1. Generation of Recombinant Constructs
The DAPK3 coding sequence was commercially synthesized (Biomatik, Canada) with a specific restriction site at two ends (BamHI/EcoRΙ). To generate a fusion protein with a traceable marker (green fluorescent protein (GFP)), the DAPK3 coding sequence was sub-cloned into the pEGFPN1vector between the cytomegalovirus (CMV) promoter and enhanced green fluorescent protein (EGFP) coding sequence. The PEGFPN1-DAPK3 (4733 base pairs) construct was verified by restriction analysis and further sequenced commercially (SeqLab, Germany).
2.2. Cell Culture and Transfection
Gastric adenocarcinoma cell line MKN-45 (Pasteur Institute of Tehran, Iran) was used for over-expressing the PEGFPN1-DAPK3 fusion protein by transfection with the purified recombinant pEGFPN1-DAPK3 plasmid, using lipofectamine 2000 reagent (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. Cells were cultured in high glucose DMEM (Biosera, USA), supplemented with 10% fetal bovine serum (Biosera, USA), glutamine (2 mM), penicillin G (100 U/mL), and streptomycin (100 mg/mL) in 5% CO2 and 37°C.
2.3. Analysis of PEGFPN1-Death-Associated Protein Kinase 3 Over-Expression in MKN-45 Cell Line Using Flow Cytometry and Western Blot
To determine the transfection efficiency and over-expression of the PEGFPN1-DAPK3 fusion protein in MKN-45 cells, the percentage of GFP positive cells was measured, using the flow cytometry technique. Briefly, 72 hours after transfection of MKN-45, cells were harvested after treatment with trypsin/EDTA and suspended in one ml of phosphate buffer saline, and directly applied to the Partec PAS flow cytometer (Partec, Germany) using a bivariate scatter plot of fluorescence versus forward scatter, by gate setting with untransfected cells. Approximately 50000 events from the transfected cell population per sample were analyzed. MKN-45 cells transfected with mock plasmid were used as the negative control. In addition, Western blot analysis was carried out to verify EGFP-DAPK3 fusion protein expression, using the goat anti-GFP antibody (ab5450, Abcam, USA). Briefly, 72 hours post-transfection MKN-45, cells were washed with phosphate buffer saline and, then, treated with cold RIPA lysis buffer in the presence of protease inhibitors cocktails (Thermo Fisher Scientific, USA). Cell lysates were collected from the culture flask and clarified by centrifugation at 13000 g and 4°C for 15 minutes. The concentration of proteins contained in clarified lysates was determined by the Peirce Rapid Gold BCA Protein Assay kit (Thermo Fisher Scientific, USA). About 25 μg of samples were loaded into 12% Tris-HCl polyacrylamide gels, and electrophoretically separated protein bands were transferred to nitrocellulose membrane (Bio-Rad, USA). After blocking the membrane with 5% skimmed milk at room temperature for 1 hour, the primary antibody (goat anti-GFP antibody) was, then, diluted in blocking buffer and incubated overnight at 4°C. Then, the membranes were washed 3 times and incubated with a peroxidase-conjugated secondary antibody (Rabbit Anti-Goat HRP (ab6741, Abcam, USA) for 1 hour. The antigen-antibody reaction was visualized, using DAB (3,3’-diaminobenzidine tetrahydrochloride (BioRad, USA).
2.4. Tetrazolium Bromide Assay
MKN-45 cells were transfected with construct pEGFPN1-DAPK3 or pEGFPN1- Mock for 12 hours and were seeded into 96-well plates (5 × 103/well). Every 24 hours until 4days after transfection, the possible inhibitory effect of DAPK3 overexpression on growth kinetic and survival rate of MKN-45 cells was evaluated by MTT assay. Briefly, after indicated times, 100 μL of MTT (5 mg.mL-1) solution (Sigma-Aldrich, Germany) was added per well, and the plates were kept at 37°C in a humidified atmosphere of 5% CO2. After 4 hours, the mediumin each well was carefully discarded and 100 μL of pure DMSO (dimethyl sulfoxide) (Sigma-Aldrich, Germany) was added to each well to dissolve insoluble formazan crystals. The absorption was read at 570 nm, using an ELISA reader. Results obtained are presented as mean ± SD of 3 independent experiments.
2.5. RNA Extraction, Complementary DNA Synthesis, and Real Time Quantitative PCR
The RNA extraction from transfected cells was performed, using the RNeasy Kit (Qiagen, USA) according to the manufacturer’s instructions. The isolated RNA was eluted in 100 μL of nuclease-free water, and the quantity of the RNA was determined spectrophotometrically by Nanodrop ND-1000 (NanoDrop Technologies, Inc, Wilmington, DE, USA). Then, complementary DNA (cDNA) was synthesized, using the Transcriptor First Strand cDNA Synthesis Kit (Roche, US). All real-time PCR reactions were performed in triplicate with 2X SYBR Premix Ex Taq technology (Takara BIO, Japan) on a Rotor-Gene Q (Qiagen, USA). Thermal cycling conditions included an initial activation step for 2 minutes at 95°C followed by 40 cycles including a denaturation step for 10 seconds and an extension step for 20 seconds at 60°C. Seven primer pairs specific for BCL2, Bax, BBC3, NOX, p62, MCL1, and Beclin1 genes were selected from a valid primer database (RTPrimerDB, Gent University). The sequences of primers used for qRT-PCR are indicated in Table 1 and the length of the PCR product is given in Table 2. For normalizing the efficiency of reverse transcription and eliminating the variations in the amount of template RNA, the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene was used as a reference to reach a standard comparison between study groups. Individual CT scores were determined as the averages of triplicate assessments. Melting curve analysis was used to assess PCR reaction. Relative transcription change was analyzed as 2-ΔΔCt, using Software Tool (REST® 2009). Results are presented as mean ± SD of 3 independent experiments (* P ≤ 0. 05 represents a significant difference from control cells).
| Gene Name | Forward Primer (5’- 3’) | Reverse Primer (5’- 3’) |
|---|---|---|
| GAPDH | TGCACCACCAACTGCTTAGC | GGCATGGACTGTGGTCATGAG |
| BCL2 | ATGTGTGTGGAGAGCGTCAACC | TGAGCAGAGTCTTCAGAGACAGCC |
| Beclin1 | TACCACAGCCCAGGCGAAAC | CCAGTGACCTTCAGTCTTCGGC |
| Bax | GATGCGTCCACCAAGAAGCT | CGGCCCCAGTTGAAGTTG |
| p62 | TTCCAGCACAGAGGAGAAGAGC | GATTCTGGCATCTGTAGGGACTG |
| MCL1 | GGACATCAAAAACGAAGACG | GCAGCTTTCTTGGTTTATGG |
| NADPH oxidase activator 1 ( NOX) | GACGTCCTGTGTGAAGTGGA | TTAGGGCTGATCTCCCTGCT |
| BCL2 binding component 3 ( BBC3) | CCTGGAGGGTCCTGTACAATCT | GCACCTAATTGGGCTCCATCT |
| Gene Name | Forward Primer Name | Reverse Primer Name |
|---|---|---|
| 1- GAPDH (87 base pair) | GAP-Fw: TGCACCACCAACTGCTTAGC | GAP-Rev: GGCATGGACTGTGGTCATGAG |
| 2- BCL2 binding component3 (88 base pair) | BBC3-Fw: CCTGGAGGGTCCTGTACAATCT | BBC3-Rev: GCACCTAATTGGGCTCCATCT |
| 3- NADPH oxidase activator 1 (153 bp) | NOX-Fw: GACGTCCTGTGTGAAGTGGA | NOX-Rev: TTAGGGCTGATCTCCCTGCT |
| 4- Myeloid cell leukemia sequence-1 (154 bp) | MCL1-Fw: GGACATCAAAAACGAAGACG | MCL1-Rev: GCAGCTTTCTTGGTTTATGG |
| 5- Beclin1 (219 bp) | Beclin-Fw: TACCACAGCCCAGGCGAAAC | Beclin-Rev: CCAGTGACCTTCAGTCTTC GGC |
| 6- Sequestosome 1 (p62) 258 base pair | P62-Fw: TTCCAGCACAGAGGAGAAGAGC | P62-Rev: GATTCTGGCATCTGTAGGGACTG |
| 7- Bax (170 bp) | Bax-Fw:GATGCGTCCACCAAGAAGCT | Bax-Rev: CGGCCCCAGTTGAAGTTG |
| 8- Bcl-2 (196 bp) | Bcl2-Fw: ATGTGTGTGGAGAGCGTCAACC | Bcl2-Rev: TGAGCAGAGTCTTCAGAGACAGCC |
| 9- HPRT1 ( 94 bp) | HPRT-Fw: TGACACTGGCAAAACAATGCA | HPRT-Rev: GGTCCTTTTCACCAGCAAGCT |
2.6. Flow-Cytometry Analysis of Cell Cycle
Briefly, 5 × 105 MKN-45 cells were seeded into 6-well plates. Then, the cells were harvested and washed twice with phosphate buffer saline (PBS) and fixed in 70% ethanol. The propidium iodide (PI) staining solution containing 50 μg/mL PI (Sigma), 10 μg/mL RNase A (Sigma), and 0.1% Triton X-100 was added to the cell pellet and incubated for 3 hours at 4°C. The population of the MKN-45 cells, which construct pEGFPN1-DAPK3, was gated by separating the GFP positive cells from negative cells. Then, the gated population was subjected to cell cycle analysis at the FL3 filter. The results were interpreted, using Flowjo software (10. 2) and the proliferation index was determined.
2.7. Assessment of Apoptosis Using Flow Cytometry
MKN-45 cells were harvested after 72 hours at a density of 3 × 105 cells, washed with PBS, and resuspended in a total volume of 100 μL of incubation buffer. Annexin-V-Flous (2 μL per sample) was added and cell suspensions were incubated for 20 minutes in the dark. Fluorescence was, then, measured, using flow cytometry. Annexin V-positive and PI-negative cells were considered to be in the early apoptotic phase and cells having positive staining both for annexin-V and PI were deemed to undergo late apoptosis.
2.8. Statistical Analysis
Experimental data were described as the mean ± SD of 3 independent assays. All tests were carried out in triplicate. An independent one-way ANOVA test and t-test were performed for comparison between groups by using SPSS 16.0 statistical software (Scarborough, Canada). All qRT-PCR results were analyzed, using REST® 2009 software. Statistically different values were defined as significant at a P value less than 0.05.
3. Results
3.1. Transfection Efficiency of the Constructed pEGFPN1 Plasmid and Verification of Death-Associated Protein Kinase 3 Expression in MKN-45 Cells
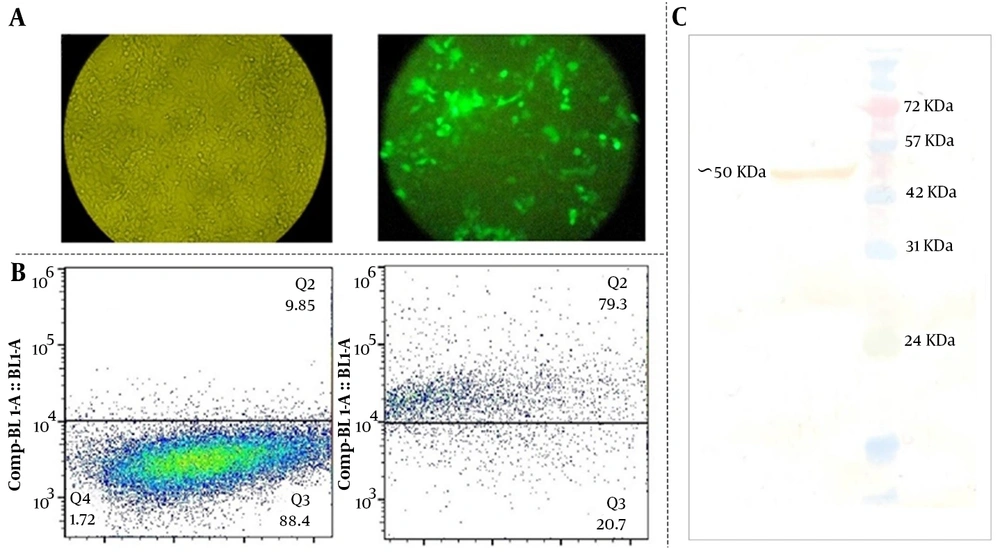
To test the effects of DAPK3 gene expression on GC, we firstly constructed non-viral vectors (pEGFPN1 and pEGFPN1–DAPK3) and then the GC-derived MKN-45 cell line was selected to be subjected to the indicated plasmids. The transfection efficiency of approximately 50000 cells from transfected cells was assessed, using both flow cytometry and fluorescence microscopy. As presented in Figure 1A, the achieving results revealed that a considerable population of the tested cells were positive for GFP. The percentage of the cells expressing GFP in the gated region was 79% after 72 hours of transfection (Figure 1B). To verify the expression of DAPK3 in MKN-45 cells thatwere transfected by pEGFPN1-DAPK3, a western blot technique was performed, using a specific anti-DAPK3 antibody. Immunoblot analysis of the lysates of DAPK3 over-expressed cells showed the recombinant protein with a size of approximately 55 kDa (Figure 1C).
Assessing the transfection efficiency and the expression of death-associated protein kinase 3 in the GC cell line. A and B, approximately 50000 cells from transfected cells were assessed the transfection efficiency was determined using both flow cytometry and fluorescence microscopy; C, Western blot of the DAPK3 protein expressed in MKN-45 cells 72 hours post-transfection by pEGFPN1-DAPK3 construct using specific polyclonal goat anti-DAPK3
3.2. The Effect of Ectopic Overexpression of Death-Associated Protein Kinase 3 on the Metabolic Activity of MKN-45 Cells
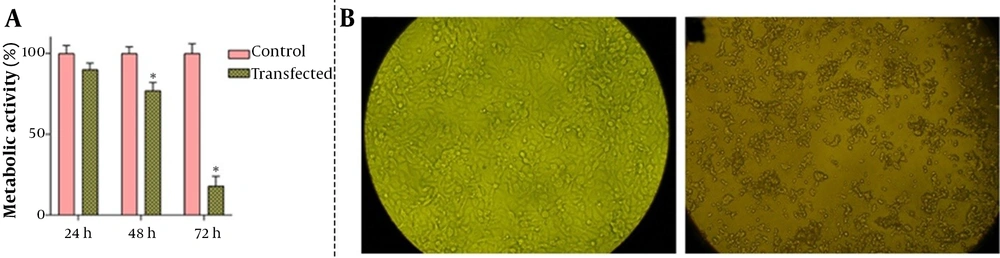
Having effectively transfected the DAPK3 gene into the cells and evaluated its expression, it was tempting to ascertain whether the elevated expression of this gene could reduce the survival of MKN-45 cells. Notably, the MTT assay with 5 × 103 cells indicated that overexpression of DAPK3 could significantly diminish metabolic activity in MKN-45 cells in a time-dependent manner (Figure 2A). It is worth mentioning that microscopic assessment during this experiment showed morphological changes in MKN-45 cells overexpressing DAPK3 with irregular membrane edges, followed by turning around and being shrunk. As represented in Figure 2B, 72 hours after transfection populations of the cells started to float in the culture medium.
The effect of death-associated protein kinase 3 (DAPK3) overexpression on metabolic activity and morphology of MKN-45 cells. A, Transfection of the DAPK3 gene into the GC cells was associated with the reduction in the metabolic activity of the cells in a time-dependent manner by MTT assay; B, Morphological changes were observed 72 hours after transfection under an inverted phase-contrast microscope with digital images captured. Values for cellular metabolic activity are given as mean ± SD of 3 independent experiments by the one-way ANOVA test. * P ≤ 0. 05 represented significant changes from the control.
3.3. The Anti-proliferative Impacts of Death-Associated Protein Kinase 3 Were Mediated Through Induction of G2/M Cell Cycle Arrest
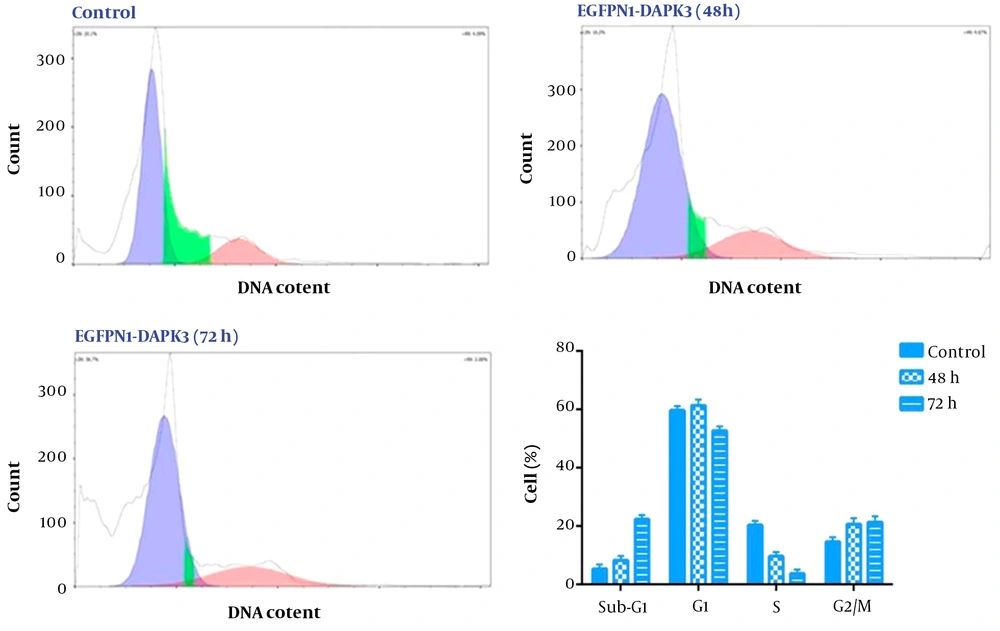
Death-associated protein kinase 3 has been suspected of the regulation of several cellular processes including cell cycle progression (14). To understand the mode of action and the anti-tumor mechanism of DAPK3 overexpression in human GCMKN-45 cells, cell cycle analysis was performed 48 hours and 72 hours after transfection of the cells (5 × 105 cells). As depicted in Figure 3, our results showed that at both time points, DAPK3 overexpression could change the distribution of the cells in different phases of the cell cycle. We found that DAPK3 not only could diminish the proportion of the cells in the S phase of the cell cycle but also could halt the transition of the cells from the G2/M phases of the cell cycle (Figure 3) proposing that the anti-proliferative impacts of DAPK3 on MKN-45 cells were mediated, at least partly, through induction of G2/M cell cycle arrest.
The effect of death-associated protein kinase 3 (DAPK3) overexpression on the distribution of MKN-45 cells in different phases of the cell cycle. Our results revealed that DAPK3 overexpression could induce G2/M arrest in MKN45 cells 72 hours after transfection. Values are given from 3 independent experiments of 5 × 103 cells.
3.4. The Effects of Death-Associated Protein Kinase 3 Overexpression on the Apoptotic Pathway
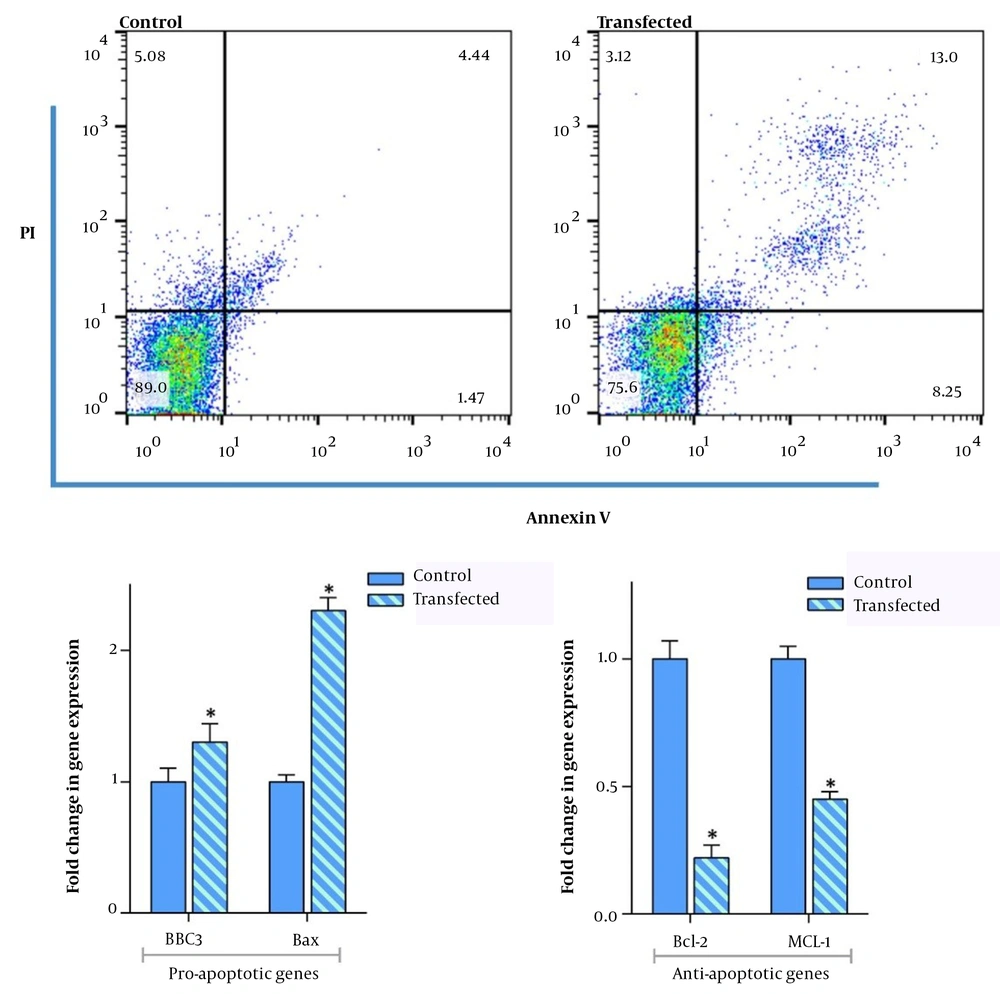
Based on the accumulating body of evidence, DAPK3 could induce apoptosis when overexpressed in mammalian cells and also can regulate this essential pathway by balancing the death-related genes (15). As represented in Figure 4, our results showed that the amounts of annexin-V positive cells were elevated in the DAPK3-overexpressing MKN-45 cells as compared to the cells in the control group. Having established its apoptotic effects, we aimed at investigating the effect of DAPK3 overexpression on apoptosis-related genes using qRT-PCR in gastric adenocarcinoma MKN-45 cell lines. For this purpose, the RNA (1 μg) was extracted 72 hours after transfection from the DAPK3 overexpressed and the untransfected control cells. As shown in Figure 4, qRT-PCR results indicate that not only does the overexpression of DAPK3 enhance the mRNA expression levels of BAX and BBC3 which play a vital role in the mitochondrial apoptotic process, but also restricted the transcription of Bcl-2 and MCL-1 anti-apoptotic genes in transfected cells as compared with the control group.
The effect of overexpression of DAPK3 on apoptosis-related genes. At a density of 3 × 105 cells and 72 hours after transfecting MKN45 cells with DAPK3, the percentage of annexin-V positive cells significantly increased as compared to untransfected control cells. The qRT-PCR analysis using 1λ cDNA also confirmed that overexpression of DAPK3 not only increased the expression of BBC3 and BAX genes but also diminished the expression levels of Bcl-2 and MCL-1. Analyzed using REST® 2009 software. Values are given as mean ± SD of 3 independent experiments and analyzed by t-test. * P ≤ 0. 05 represents significant changes from control cells.
3.5. Death-Associated Protein Kinase 3 Overexpression Could Affect Autophagy-Related Genes
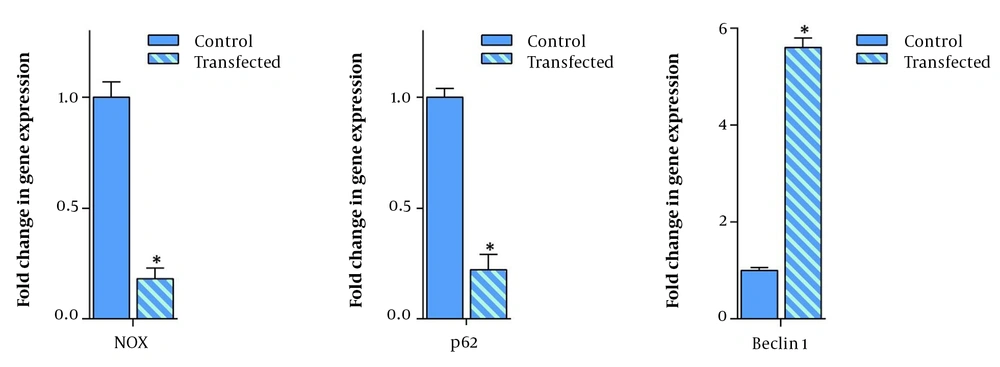
Apart from apoptosis, it has been stated that autophagy is another pathway that its regulation could be mediated at least partially via DAPK3 activation (16). Given this, it was of particular interest to examine the influence of DAPK3 overexpression on autophagy-related genes in gastric adenocarcinoma MKN-45 cell lines, using qRT-PCR. As presented in Figure 5, the resulting data clarified that the mRNA expression levels of 2 autophagy-related genes, NOX (autophagy activator) and p62 (an autophagy substrate that is used as a reporter of autophagy activity) were reduced as compared with the control group. We also found that this transfection could result in the momentous escalation of Becline1 tumor suppressor autophagy-related gene transcription in transfected MKN-45 cells as compared to the control cells.
The effect of overexpression of death-associated protein kinase 3 on autophagy-related genes. Seventy-two hours after transfecting MKN45 cells with DAPK3 and use of 1λ cDNA, qRT-PCR was performed showing a significant escalation in the expression of the Beclin1 gene. The overexpression of DAPK3 also decreased the expression of p62 and Nox genes. Values are given as mean ± SD of 3 independent experiments. * P ≤ 0. 05 represents significant changes from control cells.
4. Discussion
With fast and weight improvements in the field of molecular biology and biotechnology, a stunning window of hope has been opened in front of patients with different malignancies and their physicians. In this regard, the comprehension of disease mechanisms at the molecular level together with the recognition of myriad onco- and tumor suppressor genes have provided an extensive potential for gene therapy so far (17). Given the fact that plasmid-mediated gene therapy has been able to achieve beneficial consequences in numerous in-vivo and in-vitro studies, intense interest has been devoted to plasmid vectors, especially the non-viral ones which possess various benefits over viral ones in respect to their safety and user-friendliness (18). Many clinical studies have now been performed, using non-viral technology by introducing genes into the genome of cancerous cells and producing adequate amounts of the gene product to induce obligatory cell death mediated by overexpression of a definite anti-cancer gene.
Death-associated protein kinase 3, a member of the DAPK serine/threonine-protein kinase family, is known to possess cancer-specific cytotoxicity effects and is also stated to serve as a tumor suppressor through regulation of apoptosis, proliferation, and starvation-induced autophagy (19). In the present study, we planned to examine whether constructing a non-viral plasmid encoding DAPK3 that can efficiently carry this gene into a human gastric adenocarcinoma MKN-45 cell line could be an effective strategy to reduce the survival of the neoplastic cells. The results of our study revealed that the overexpression of DAPK3, as approved by western blot analysis, not only decreased metabolic activity in transfected MKN-45 cells in a time-dependent manner but also increased the percentage of the cells in the G2/M phase, suggesting that the anti-proliferative effect of the gene is mediated, at least partially, through the induction of G2/M arrest in GC cells. Our results were inconsistent with the results of the study of Kake et al. who demonstrated that DAPK3 knock down increased proliferation, migration, and invasion of A549 cells (11). It is been proposed that the members of the DAPKs family can be involved in cancer cells in 2 ways. They not only can act through the mTORC pathway but also can act through Becklin1 (16, 20). In this way, by activating the Becklin1 factor, they activate the autophagy pathway, whereas, via the mTORC pathway, they inhibit autophagy, the inconsistency of our results with the results of Kake et al. (11) can be explained by the possible dual role of this protein in cancer cells (20, 21). On the other hand, our results were in agreement with a previous study showing that escalation in the mRNA expression levels of DAPK3 could significantly diminish cell proliferation in a dose-dependent manner in PC-3 prostate carcinoma cell line (11).
It has been demonstrated that DAPK3 could induce apoptosis through either caspase-dependent or -independent signaling (13, 22). This enzyme contains an N-terminal kinase domain, which plays a vital role in the induction of cell death (23). Of note, the molecular analysis demonstrated that overexpression of DAPK3, on one hand, increased the mRNA expression levels of BAX and BBC3 pro-apoptotic genes, and on the other hand, decreased the transcription of Bcl-2 and MCL-1 anti-apoptotic genes, which all were in agreement with the elevated cell population in the sub-G1 phase of the cell cycle. In harmony, Kawai et al. reported that DAPK3 overexpression positively regulates apoptosis and suggested that the catalytic activity of this enzyme is associated with the induction of programmed cell death (24). In another study conducted by Brognard et al., it has also been stated that overexpressed DAPK3 caused a > 2-fold increase in apoptosis (25). Autophagy, an evolutionarily conserved intracellular degradation system (26), is a process that influences most cancers with 2 different consequences; either it could restrict tumorigenesis or lead to cancer progression by increasing cell survival (27, 28). Noteworthy, we demonstrated that DAPK3 overexpression suppressed the expression of NOX (autophagy activator) and p62 (an autophagy substrate that is used as a reporter of autophagy activity) genes in the transfected MKN-45 cell line. In addition, the results of our experiments showed that the overexpression of DAPK3 increased transcription of Beclin1, which acts as a tumor suppressor and is an essential mediator of autophagy. The correlation between that DAPK3 overexpression and induction of autophagy-related cell death has been reported in several adenocarcinoma cells (16, 29, 30). In this vein, Li et al. also shed light on the ability of DAPK3 in phosphorylating ULK1, a well-known enzyme that could activate autophagy in response to amino acid withdrawal, suggesting that the tumor suppressor property of this molecule might be mediated through stimulation of autophagy flux (20). In conclusion, our experimental investigation indicated that the overexpression of the DAPK3 gene can lead to cell death by both inducing apoptosis and autophagy pathways in gastric adenocarcinoma. This anti-cancer activity may describe a hopeful strategy in the application of novel gene therapy for the treatment of gastric adenocarcinoma; however, further research is required to examine the clinical effectiveness of this strategy in GC treatment.