1. Context
The innate immune system, or nonspecific immune system, is considered the first line of host defense, which is comprised of cells and mechanisms capable of mediating the earliest interactions with pathogens (1). The innate immune system relies on “pattern recognition” that can be recognized by pattern recognition receptors (PRRs); PRRs recognize pathogen-associated molecular patterns (PAMPs) (2). Toll-like receptors (TLRs) are a family of PRRs, which can discriminate molecules from pathogens and antigens from host molecules, collectively referred to as pathogen-associated molecular patterns (PAMPs) (3). A variety of ligands, including different types of organisms or structures can bind to and activate TLRs (4, 5). TLRs are expressed by different types of leukocytes. A microRNA (miRNA) is a non-coding RNA molecule, which can regulate gene expression (6). Recent studies have discovered a relation between miRNAs and immune systems in cancer (7, 8). Lymphocytic leukemia was the first disease, which was associated with miRNA deregulation. MiRNAs were demonstrated to play the important roles in cancer and, accordingly, are sometimes referred to as “oncomirs” (9). Melanoma, the most dangerous type of skin cancer, is a type of these cancers, which can originate from the pigment-containing cells or melanocytes (10). Melanomas can occur in the skin mouth, intestines, or eyes. Melanoma usually occurs on the legs and back in women and men, respectively (11).
2. Evidence Acquisition
We searched PubMed, Medline, Scopus and Google Scholar databases for the studies published from January, 1998 to October, 2017 with the following keywords: “TLRS”, “MicroRNA”, “cancer”, “HMGB1”, and “Melanoma”.
3. Results
3.1. MicroRNAs in Melanoma
The expression level of miRNAs is different in tumor cells; some miRNAs regulate onco proteins that can down regulate malignant cell transformation (12). Table 1 summarizes the main functions of miRNAs involved in melanoma (9, 12-14).
| MiRNA | Source | Association | Ref |
|---|---|---|---|
| Let7 a | Melanocyte | Migration | (11) |
| Let7 b | Melanocyte/Metastasis | Proliferation/Migration | (11) |
| mir-155 | Melanocyte | Proliferation | (15) |
| mir-324-5p | Melanocyte | Proliferation | (16) |
| mir-34a | Melanocyte/Primary melanoma | Proliferation/Migration | (17) |
| mir-34b/c | Primary melanoma | Migration | (9) |
| mir-106a | Primary melanoma/Metastasis | Proliferation/Migration | (18) |
| mir-126 | Primary melanoma | Proliferation/Migration | (19) |
| mir-133a | Melanocyte/Primary melanoma/ Metastasis | Proliferation/Migration | (19) |
| mir-141 | Primary melanoma | Proliferation/Migration | (20) |
| mir-145 | Primary melanoma | Proliferation | (21) |
| mir-15b | Primary melanoma | Proliferation | (19) |
| mir-200c | Primary melanoma | Proliferation/Migration | (19) |
| mir-210 | Primary melanoma | Proliferation | (13) |
| mir-27b | Primary melanoma | Proliferation/Migration | (22) |
| mir-199a | Primary melanoma/ Metastasis | Migration | (23) |
| mir-92a | Metastasis | Proliferation | (24) |
| mir-182 | Metastasis | Migration | (25) |
| mir-221/222 | Metastasis | Proliferation/Differentiation | (18) |
| mir-18 | Metastasis | Proliferation | (17) |
| mir-19a | Metastasis | Proliferation | (26) |
MicroRNAs Involved in Melanoma
3.2. High-Mobility Groups and Their Functions
High-mobility group (HMG) proteins are nuclear proteins, which regulate various DNA-related activities (27). HMGs bind to DNA and chromatin, and induce both short- and long-range changes in the structure of their binding sites (28). They can also affect numerous regulatory molecules such as p53 (18). HMG proteins play an important role in human disorders (29). Anti-bodies directed against HMG proteins were found in patients with autoimmune diseases and cancers (26).
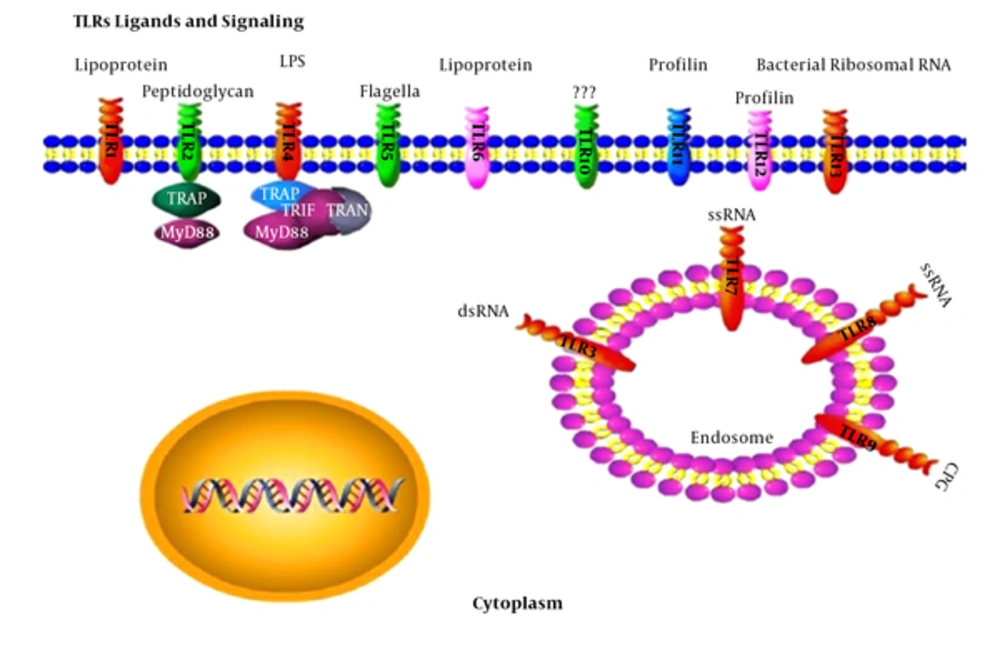
3.3. TLR Ligands and Signaling
TLRs function as dimmers, such as TLR2 and TLR6 that are hetero dimers. Each dimer contains different ligand specificity (17). TLR activity depends on their ligand sensitivity, such as TLR4 that can recognize bacterial lipopolysaccharide (LPS) (16). Activation of these receptors produces inflammatory cytokines, such as IL-6 and INF gamma (15). Adapter proteins and kinases that mediate TLR signaling have also been targeted. Some adaptor molecules, such as Myeloid differentiation primary response gene 88 (myD88) and TRIF can serve as signaling molecules of TLRs (17, 20). TLRs have been supposed to bind to the host molecules, including fibrinogen (involved in blood clotting), heat shock proteins (HSPs), and HMGB1. However, under the inflammatory and autoimmune conditions, TLRs can form a complex with endogenous proteins (30). Figure 1 summarizes TLR ligands and signaling.
3.4. MicroRNAs and TLRs
MicroRNAs play a role in up regulation mechanisms of TLR ligands, some of which directly target components of the TLR signaling pathways (21). TLR signaling must be strictly regulated by immune responses against pathogens. Some of the miRNAs, such as miR-155, miR-Let7 a/b, and miR-146a, were confirmed to be key TLR signaling molecules (31).
3.5. Let7 Relation with TLRS and HMG Proteins
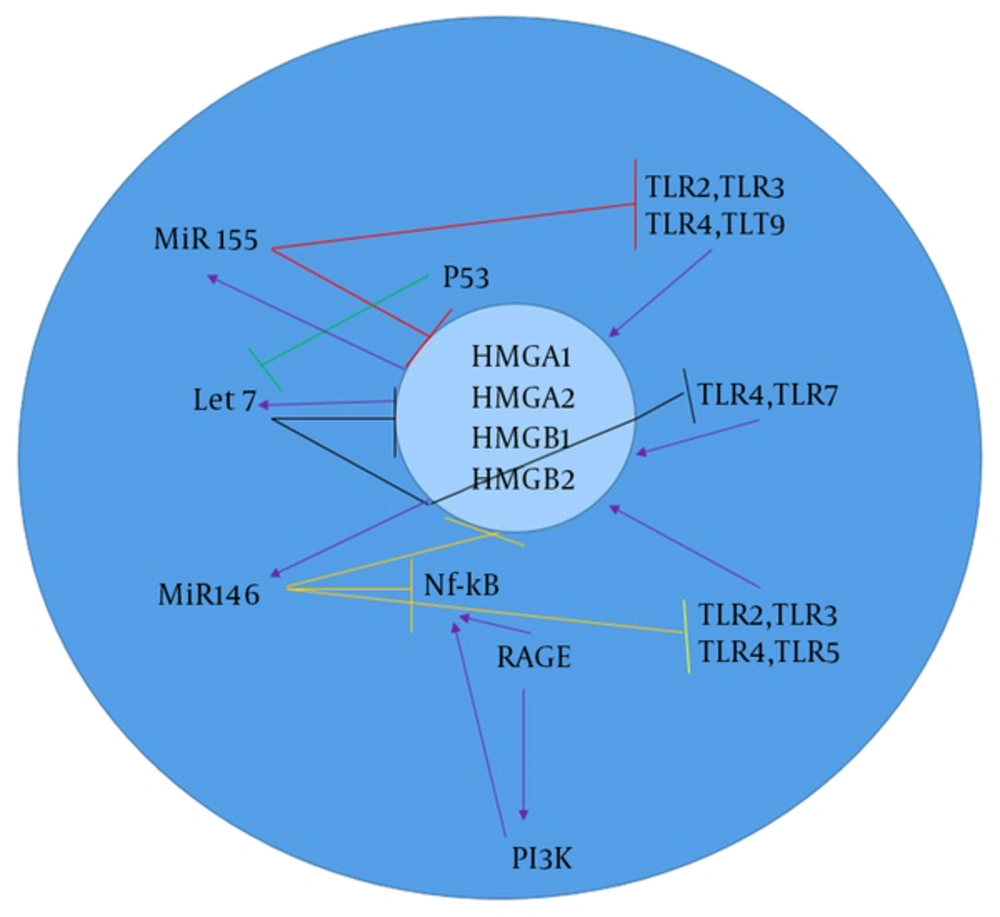
Members of the Let-7 miRNA family have an important role in suppressing melanoma tumor growth. Thirteen different members of the let-7 family, including let-7a-1, let-7a-2, let-7a-3, let-7b, let-7c, let-7d, let-7e, let-7f-1, let-7f-2, let-7g, let-7i, miR-98, and miR-202 were discovered in human beings (14). Let-7 activates TLR-7 and causes degeneration in cancer tissues (32). The main function of Let7 is to regulate the gene expression of CNS and cancer, which can also be expressed in macrophages (Figure 2) (24). Let7 can bind directly to TLR-7 and TLR-8 in macrophages and enhance the expression of pro-inflammatory cytokines, such as TNF-α and IL-6 (33).
Therefore, miRNAs are the agonists of TLR7 that are important molecules in the NF-κB signaling pathway and secretion of pro-inflammatory cytokines (20). Recent studies have shown that let-7b, along with TLR4, is involved in the inflammation and immune responses (25). Let-7e decreases the expression of TLR4 on the surface of macrophages, and let-7i is an antagonist of TLR4. Let-7 negatively regulates TLR4 activity, a specific receptor of LPS; that is, TLR4 is down regulated by let-7 (25). Let-7 is a main direct regulator of RAS expression in human cells. The 3 RAS genes, including K-, N-, and H-ras have the predicted let-7 binding sequences in their 3'UTRs (19). HMGs have 3 superfamilies, including HMGA, HMGB, and HMGN (34). HMGA1 can regulate tumor progression in a stem cell-like state (34). HMGA1 can bind to the signal transducer and activator of transcription 3 (STAT3) and can be up regulated in melanoma cell lines. HMGA1 and HMGA2 genes were found to be expressed in several cancer types, but not in adult healthy tissues. HMGA2, as a protein and competing endogenous RNA (ceRNA), was shown to modify gene expression by activation of let7 (35). HMGA1 and HMGA2 are the regulators of let-7 in melanoma so that HMGA1 and HMGA2 are negatively regulated by let7. Let-7 directly inhibits HMGA2 with binding to its 3’UTR (36). The removal of let-7 binding site through 3’UTR deletion can lead to increased expression of HMGA2 as well as tumor formation (36). Then, let-7a can indirectly block STAT3 transcription (36). STAT3 activation is necessary for NF-kB activation. HMGB1 is another superfamily of HMG proteins, which stimulates DNA binding of several steroid receptors, including let-7. HMGB1 is not a direct let-7 target, and the expression of let-7 is modulated by HMGA1 (36). Recent studies showed that the tumor suppressor p53 can down regulate the activity of the HMGB1 promoter and decrease let-7a and let-7b expression in human melanoma (36, 37).
3.6. MiR-155 Relation with TLRs and HMG Groups
MiR-155 is encoded by the MIR155 host gene or MiR155HG, which have physiological and pathological processes (38). MiR-155 regulates TLR signaling by targeting MyD88 as a negative regulator (38). MiR-155 is involved in immunity through modulating the humoral and innate immunity (24). Recent studies showed that B-cells, T-cells, and antigen-presenting cells (APCs), such as macrophages and dendritic cells have a high expression level of MiR-155 that is essential for proper of T-cell development and maturation (37). In macrophages, miR-155 is up regulated in response to the TLR3 ligand and CpG (TLR-9) (23). MiR-155 is also induced by TLR signaling (39). MiR-155 is up regulated by HMGB1 in DAMP-derived molecules. In addition, HMGB1 induces inflammatory effects, which are blocked by MiR155 in melanoma (40).
3.7. The associationofmir-146 with TLRs and HMG Groups
MiR-146 and miR-146b genes, whose sequences differ only in 2 nucleotides, are located on human chromosome 5 and 10, respectively (22). MiR-146a negatively regulates NF-κB activation. In addition, the activation of TLR4 and molecular cascade leads to decreased NF-κB activation and nuclear translocation that can induce pri-miR-146a (41). TLR signaling manages the regulation of miR-155, miR-146, and miR-132. MiR-146 negatively regulates the expression of Interleukin-1 receptor-associated kinase1 (IRAK1), IRAK2, and TNF receptor-associated factors 6 (TRAF6) (42). The increased level of miR-146a/b in melanoma significantly down regulates the expression of IRAK1 and TRAF6, and negatively regulates NF-Kb activation. MiR-146a has different effects on growth and metastases of melanoma. Th1 cells exhibit a more expression level of MiR-146a than Th2 or naïve T-cells (42). MiR-146b promotes myogenic differentiation and modulates multiple genes, such as HMGA2 and HMGA1 targets in muscle cells. The inhibition of mir-146b can down regulate HMGA2 during differentiation (42).
4. Conclusions
The detection of miRNAs has added a new layer of cancer therapy and will continue to increase our knowledge in this regulation. TLRs are the main key of smooth initiation, resolution, and control of inflammatory signals. The present study showed the important relationship between TLRs and miRNAs in targeting HMGs. The important point in this field is that miRNA targeted in the immune defense can regulate inflammatory cytokines and alter cancerous phenotypes. Most importantly, MiR-146a, miR-155, and let7 can decrease TLR function and HMG targeting, but induce the miRNA secretion in melanoma.