1. Background
Brain glioma tumors are among the most prevalent malignancies, and early management is important in patients’ survival (1, 2). Gliomas are classified into 4 grades based on the histopathologic characteristics (3). The World Health Organization (WHO) has categorized gliomas into 2 main groups of high and low grades, each subdivided into grades 1 to 4, depending on the invasiveness (4). Grade 4 gliomas, termed glioblastoma multiform, are the most invasive type with the lowest survival rate (4). Thus, making a correct diagnosis of the tumor’s grade is the critical element of the effective treatment plan (5, 6).
Radiologic images obtained via MRI, using T1, T1c, T2, or fluid-attenuated inversion recovery (FLAIR) methods, provide the standard information and assist in the clinical decisions for an effective treatment plan (7). Grading gliomas by histopathology is costly and time-consuming. In recent years, modern non-invasive, rapid, safe, and inexpensive methods of making an efficient diagnosis, by combining artificial intelligence (AI) algorithms, are becoming increasingly popular in the management of brain tumors, including gliomas. Specifically, using the AI approach is a prudent step, since making the diagnostic and treatment decisions based on MRI scans alone may be difficult and associated with irreversible errors (8). One popular AI approach is machine learning, which uses the known patterns of human brain data processing for solving complex problems (9, 10). Also, other AI components, i.e., deep learning and convolutional neural networks (CNN) combined with sequences from MRI have shown promising outcomes, thus making significant contributions to the complex task of pathological grading and classification of gliomas (9, 10).
Even though various AI methods have been significantly helpful to the practice of diagnostic radiology, future advancements are needed to improve the lesion detection, segmentation, and classification of brain tumors including gliomas (11). This is a challenging goal when it comes to choosing appropriate deep learning methods to detect and classify gliomas that may occur in the brain (12). Both machine learning and deep learning offer great potentials to contribute advances to the radiologic diagnosis of gliomas (8, 13-15). To date; however, today there are certain questions on the role of AI in the grading and classification of human gliomas that remain unanswered, which are the impetus behind our planning for and undertaking this study.
2. Objectives
This study was conducted to develop a transfer learning model to efficiently and accurately grade glioma tumor sequences, using established data derived from the MRI scans of patients with gliomas.
3. Methods
3.1. Study Design
The study design was approved by the Ethics Committee, Dept. of Information Technology Management, Faculty of Management and Economics, Science and Research Branch, Islamic Azad University, Tehran, Iran (Ethics Code: IR.IAU.SRB.REC.1399.052). This study only involved in the collection and analysis of data from an original dataset entitled BraTS-2019, which had been developed and validated in 2019 by the Center for Biomedical Image Computing & Analytics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA. All of the human data that were derived from the original dataset were anonymous and kept strictly confidential during and after the completion of this study.
3.2. Literature Review
To select an appropriate deep learning method, we conducted a review of credible online literature. Initially, 190 published articles were identified based on their titles and keywords. Upon a careful review of the articles, 35 of them were found to be appropriate as the criteria for choosing a deep learning method.
3.3. Training the Model
The model was trained with the BraTS-2019 dataset, originally derived from the standard MRI scans from 335 patients with brain gliomas. Of the 26,904 images in the dataset, 20% were used for the validation with the other 80% for the training of the proposed model. The anonymous images were divided into two groups, consisting of 259 high-grade (HGG) and 76 low-grade gliomas (LGG). To create a balance between the two groups of images, similar numbers of 2-D sequences from the HGG (n = 13,233) and LGG (n = 13,671) were used to train our proposed model.
3.4. Deep Learning Models
Deep learning models are normally constructed by two approaches:
a) A model with all of the associated layers developed by the researchers. This was not our choice due to its high cost and being time-consuming.
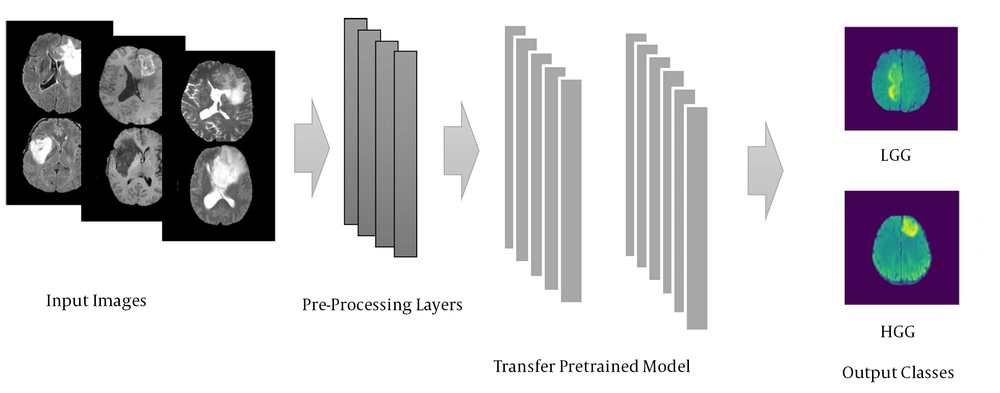
b) A model known as transfer learning is used with the layers adopted from other pretrained models. Such a model is developed by well-known suppliers and is currently popular in AI applications. We used one of the small models from this AI family, called EfficientNetB0, for the current study. Figure 1 illustrates the schematic view of the proposed transfer learning model, and the processing of input images through the initial and advanced layers. After process, the input MRI scans were graded and categorized into two groups of either LGG or HGG images.
3.5. Image Data Analyses
To include the images and edit them before training the model, the 3-D images were converted to 2-D ones by a data enhancement technique, using random flip and rotation methods, based on specific codes and parameters. Then, we designed the network layers, and accurately refined the parameters for transfer learning. Subsequently, we utilized a pretrained convolutional neural network (EfficientNetB0) as our base model (16), which is the most recent and efficient transfer learning model for image classification. Upon training the model, we analyzed the results for accuracy, sensitivity, specificity, and precision. As indicated earlier, 20% of the entire images (n = 26,904) were used for validation, with 80% remaining used for the model training.
3.6. Normal vs. Abnormal Images
The proposed model did not read normal images due to hardware and software limitations. Otherwise, it would require additional training before it could differentiate normal from abnormal ones. Thus, we excluded the processing of normal images.
3.7. Experimental Setup
For each patient, there were 5 brain tumor sequences available in the original dataset. We used 3 of the sequences; i.e., T1ce, T2, and fluid-attenuated inversion recovery (FLAIR). The latter is an MRI sequence with the inversion recovery set to null fluids. Also, one MRI sequence was assigned as the ground truth. The format for all sequences was Neuroimaging Informatics Technology Initiative (NIFTI), with the length, width, and slice numbers being 240, 240, and 155, respectively. The image size for inclusion in the proposed model was 240 × 240, with the batch size being 32. Details are shown in Table 1.
Abbreviations: HGG, high-grade glioma; LGG, low-grade glioma.
a Number of patients in the original dataset.
b Extracted data from the original dataset.
Also, we utilized Mango software for the initial image visualization. The model was implemented in Python programming language, using Keras library and Tensorflow backend. Also, we took advantage of the SimpleITK library to read the MRI scans, and the Matplotlib library to illustrate images. Further, we processed each image to 50 epochs only, due to hardware limitations (Table 2).
a Momentum 1.
b Momentum 2.
c Number of rounds the dataset training completed to process each image.
3.8. Image Reading Process
Finally, we employed Google Colab to execute the image readings. This step provided an environment for writing and implementing the Python codes based on Cloud, and enabled access to GPU and a variety of P100s, P4s, T4s, and Nvidia K80s.
4. Results
Based on the methods, the proposed model was capable of predicting the tumor grades for the images at high accuracy. Details of the results via the model and its performance are presented below:
4.1. Validity and Versatility
Assuming that employing multiple sequences would provide more useful data for grading than using a single sequence, the obtained data to initiate the grading were based on T1ce, T2, and FLAIR sequences. Since we used the T1ce sequences, the T1 counterparts were not used, otherwise, additional hardware was needed, which we lacked. Table 1 presents the number of qualified images used for grading the HGG and LGG images. Based on the codes and parameters, and by image augmentation, we correctly included the MRI scans in the proposed model (Table 2).
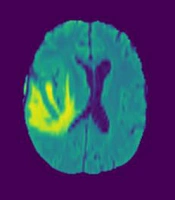
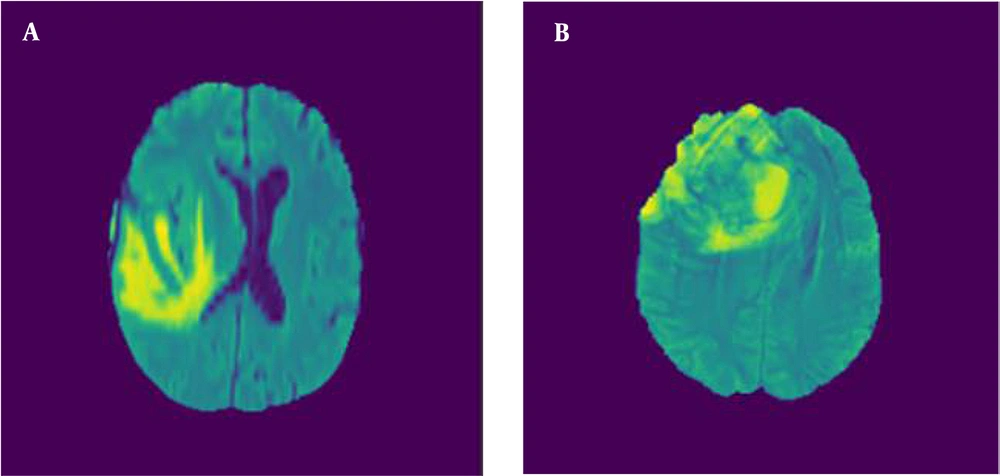
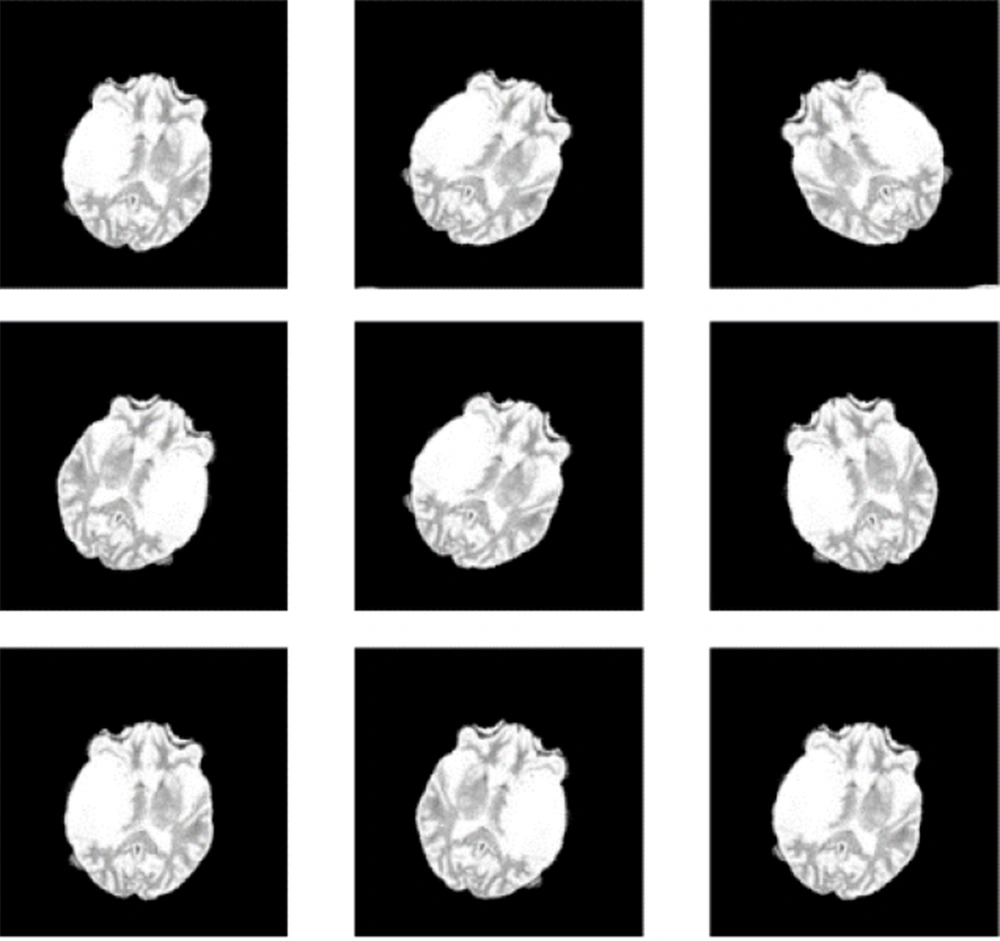
Figure 2 illustrates the axial MRI views of LGG and HGG taken at axial angles, based on FLAIR sequences. Figure 3 shows a series of altered MRI scans of gliomas with the augmentation technique after being processed by the proposed model.
4.2. Performance Assessment
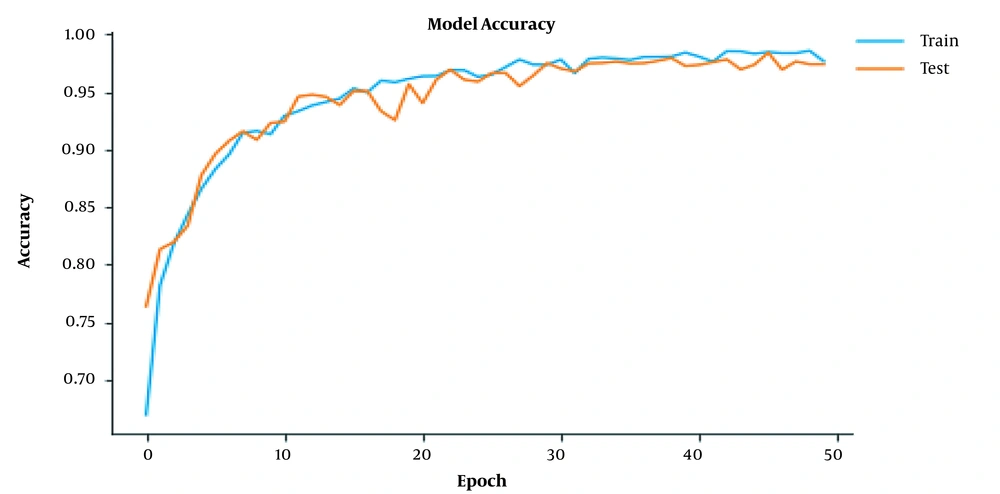
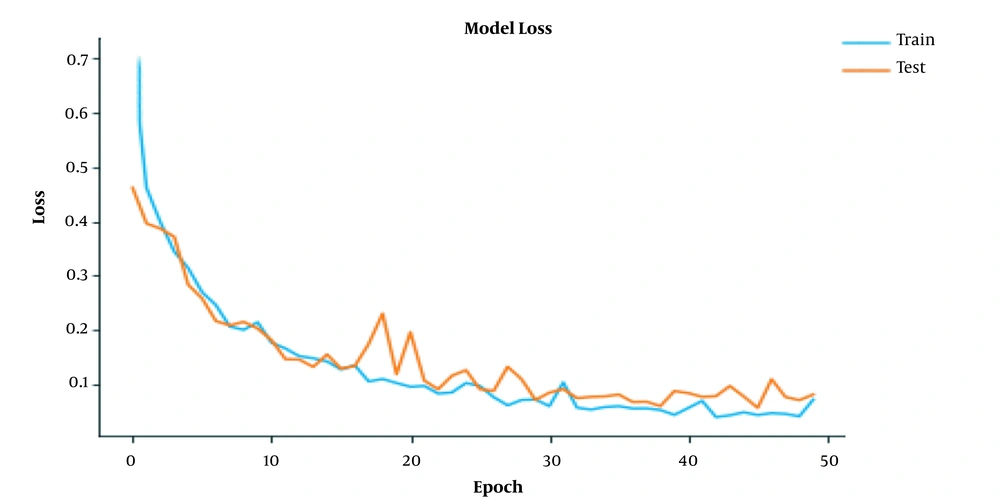
To assess whether the model was overfitting, we used a cross-validation technique, for which the images were divided into separate training and validation sets. Figures 4 and 5 represent the performance of the model during the training processes. The proposed model was ready for grading tumor sequences after 50 epochs of training. As the training epochs increased, the accuracy of model also improved further (Figure 4). Therefore, both the training and test curves progressed together consistently at a similar rate. Also, by increasing the number of training epochs, the network loss declined exponentially, so both the training and test plots approached zero level at similar rates (Figure 5).
Upon developing and training, the model was capable of predicting the grades and classes of tumor sequences that had not been seen before or during the dataset training. The architecture, image dimension base datasets, and the accuracy of the model for classifying HGG and LGG images versus those achieved by seven earlier models between 2018 and 2020 have been compared, as are shown in Table 3.
| Study | Model’s Architecture | Dimension | Dataset | Accuracy |
|---|---|---|---|---|
| (17) | Transfer Learning (VGG-16) | 2D | TCIA | 0.9500 |
| (18) | CNN | 3D | TCIA | 0.9125 |
| In-house | 0.9196 | |||
| (19) | CNN | 2D | Brats-2013 | 0.9943 |
| BraTs-2014 | 0.9538 | |||
| BraTs-2015 | 0.9978 | |||
| BraTs-2016 | 0.9569 | |||
| BraTs-2017 | 0.9778 | |||
| ISLES-2015 | 0.9227 | |||
| (20) | Transfer Learning (AlexNet) | 2D | In-house | 0.8550 |
| Transfer Learning (GoogLeNet) | 0.9090 | |||
| Pre-trained AlexNet | 0.9270 | |||
| Pre-trained (GoogLeNet) | 0.9450 | |||
| (21) | CNN (2D Mask R-CNN) | 2D | BraTs-2018 TCIA | 0.9630 |
| CNN(3DConvNet) | 3D | 0.9710 | ||
| (22) | Multi-stream CNN | 2D | BraTs-2017 | 0.9087 |
| (23) | 3D CNN | 3D | BraTs-2018 | 0.9649 |
| Proposed Model | Transfer learning(EfficientNetB0) | 2D | BraTS-2019 | 0.9887 |
The accuracy of the old models was the reason behind comparing their performance and accuracy with those of the proposed model. The performance and accuracy of the earlier models fell between 0.8550 and 0.9978%. The accuracy of our EfficientNetB0 model was 0.9887. Our proposed model was also evaluated for its training capacity and validity of the graded images (Table 4).
| Performance | Training Set | Validation Set |
|---|---|---|
| Accuracy | 0.9905 | 0.9887 |
| Precision | 0.9915 | 0.9898 |
| Sensitivity | 0.9934 | 0.9886 |
| Specificity | 0.9920 | 0.9879 |
5. Discussion
The proposed model accurately classified the HGG and LGG images compared to those achieved in earlier studies (Table 3), which justifies our decision to choose the results achieved in the earlier studies versus those of our model. Specifically, our model was able to reliably classify the images with an accuracy of almost 99%, whether they belonged to the LGG or HGG group. The model performance was consistent with the acceptable validity level suggested by numerous earlier studies on grade-prediction and classification models (3, 8, 9, 12, 13, 17, 18, 24). One of the weaknesses of machine learning is the extraction and classification of specific data from the images it reads. The capability of deep learning has resolved this issue, hence the reason for its popularity. Also, the large number of images used in the deep learning dataset provides for better training and prevents the overfitting the model.
5.1. Validity and Versatility
Our decision to convert 3-D to 2-D images was justified by the fact that one 3-D image can be segmented into 150 slices of 2-D images, before being used for model training. The basis for feeding a total of 26,904 qualified 2-D images of either HGG or LGG gliomas into the model was that the more images used for training, the more efficient the model became. The use of T1ce, T2, and FLAIR sequences to develop the model, made it more versatile for reading images from MRI sequences compared to using a single MRI sequence. Also, based on its excellent performance, the proposed model is capable of being further enhanced for its tumor grading validity. This may be further improved by feeding it with additional images or training it with other advanced datasets.
5.2. Comparison of Deep Learning Models
The proposed model is not only comparable to older models (19, 22) in terms of validity, accuracy, and performance but may also be a better tool to assist in the grading of gliomas. The model detects extra image details since it was trained on greater MRI sequences. A recent study (21) developed a diagnostic model based on the BraTS-2018 dataset and TCIA, using both 2-D and 3-D CNN models, and slightly achieved better results from the 3-D than the 2-D images. In a similar study (24), the 2-D model achieved an almost 99% validity for grading various grades of gliomas. An earlier study (20) used local datasets and the transfer learning method to train the pretrained networks (GoogleNet & AlexNet) and compared their predictive and diagnostic performances. This study achieved better results from GoogleNet than from AlexNet.
5.3. Image Classification, Segmentation & Grading
The use of 2-D images in our study was justified since enough 3-D images were not available to us. Thus, we expanded the dataset of the study by converting the limited 3-D images into 2-D ones. Further, using 3-D images to train a model requires a large memory, which limits the network’s resolution and lowers its representation capability (7, 25). Also, using 3-D images increases the computational cost and requires GPU memory (19). A recent study (18) developed a machine learning model for diagnosing benign from malignant gliomas, using a pretrained CNN model and validating on multiple versions of BraTS datasets.
5.4. The 2D vs. 3D Advantages
Our main reason behind using the 2-D images was to develop a non-invasive and highly accurate transfer learning model and to validly diagnose the grades of 2-D MR glioma images. This approach allowed us to generate a large number of 2-D images from a single 3-D image, which improved the prediction accuracy of the model to almost 99%. Despite previous research, the role of the deep learning method in the improvement of tumor grading needs further exploration (8). A recent study has achieved excellent validity, accuracy, and specificity for this important purpose, using a transfer learning model that was pretrained with a CNN model based on the TCIA dataset (18).
5.5. Limitations
This study had limited access to the hardware and software required for processing 3-D images, therefore, it was more practical for us to convert the 3-D images into 2-D ones.
5.6. Conclusions
The proposed model developed by this study can classify and grade glioma MRI sequences at high accuracy, validity, and specificity. Also, its performance is consistent with those suggested by numerous earlier studies on tumor grading and classification. In the absence of hardware and technical limitations, it is plausible to achieve future diagnostic models applicable to MRI sequences with accuracy and performance superior to the currently existing deep learning AI models. Presumably, using the largest model of the EfficientNet family (i.e., EfficientNetB7), which has ample trainable capacity, is likely to provide researchers with the means of developing future AI diagnostic models with superior accuracy, performance, and versatility.