1. Background
Oral squamous cell carcinoma (OSCC) is one of the most common cancers and accounts for more than 90 oral malignancies (1, 2). The mortality rate from OSCC is so high that despite the existing treatment methods, the 5-year survival is still low (1). Oral squamous cell carcinoma is, therefore, an important health problem in the global community (2). Oral squamous cell carcinoma occurs in a multi-step process, from the step-by-step accumulation of genetic mutations, which is morphologically known as a precancerous lesion (3). But, mere microscope-based clinical signs cannot determine the progression of a precancerous lesion to malignancy. Therefore, it seems necessary to study and find molecular markers that can represent such a feature (3).
One of the leading causes of cancer is long non-coding RNAs (lncRNAs) (4). These elements are more than 200 nucleotides in length and account for 80% of all transcripts in cells (5). Lnc-RNAs are expressed in the mammalian genome and are the main regulators of embryonic pluripotency, differentiation, developmental transitions, and histone modification (6-8). Studies have shown that lncRNAs, as a regulator, play an important role in the development of cancer and play an important role in this disease by enhancing tumor cell proliferation, inducing angiogenesis, metastasis, invasion, and preventing growth suppression (9, 10).
The biomarker role of gastric carcinoma highly expressed transcript 1 (GHET1) lncRNA in a variety of cancers such as prostate cancer (11), non-small cell lung cancer (NSCLC) (12), hepatocellular carcinoma (HCC) (13) has been reported and a significant relationship between this lncRNA with clinicopathological variables such as tumor stage and its differentiation have been shown (13, 14). LncRNA GHET1 appears to play a regulatory role in increasing cancer cell proliferation by inhibiting apoptosis (11). In hepatocellular carcinoma, the lncRNA GHET1 is induced by H3K27 acetylation and stimulate cell carcinogenesis through activating ATF1 (15). Gastric carcinoma highly expressed transcript 1 lncRNA knockdown reduces cell-cycle progression and invasion (16). Gastric carcinoma highly expressed transcript 1 lncRNA knockdown suppresses colorectal cancer cell growth and invasion (17). Overexpression of the lncRNA GHET1 enhances multidrug resistance in gastric cancer cells. While various research reporting a link between lncRNA GHET1 and malignancies, no coherent conclusion on the prognosis usefulness of lncRNA GHET1 in patients with cancer has been reached due to diverse results and small sample sizes in each investigation (12, 14-18).
ZXF2 lncRNA is located on 8q24.2 and, recently, overexpression of ZXF2 lncRNA in lung adenocarcinoma has been reported (19). Examining the relationship between the expression of this element and the outcome in patients with lung adenocarcinoma showed that upregulation of ZXF2 lncRNA is linked to increased lymph node metastases and a poor prognostic (19). It was expressed 8.879-fold more frequently in lung adenocarcinoma specimens than in non-cancerous tissues. However, the role of ZXF2 lncRNA in other cancers has not been reported. Further investigation into the molecular mechanisms underlying ZXF2-mediated tumor progression revealed that siRNA-mediated suppression of ZXF2 resulted in cell cycle arrest and inhibition of cell proliferation, migration, and invasion by regulating c-Myc, a strong proto-oncogene found nearby to ZXF2 in chromosome loci 8q24.2, a prevalent location for multiple tumors (19).
2. Objectives
Given that no study has been conducted on the roles of lncRNA GHET1 and lncRNA ZXF2 in OSCC and their relationship with clinicopathological variables, therefore, the present study aimed at evaluating the expressions of lncRNA GHET1 and lncRNA ZXF2 in OSCC patients for early detection of it.
3. Methods
3.1. Patients and Tissue Sampling
Thirty OSCC samples and normal tissues from the tumor's margin were separately produced from patients who underwent oral and maxillofacial surgery at Imam Khomeini Hospital in Tehran, Iran, under the observation and permission of a specialist. The present study was performed with the approval of the Medical Research Ethics Committee (IR.IAU.PS.REC.1399.029) of the hospital and with the written consent of all patients, as well as following the Helsinki Statement. Patients receiving chemotherapy or radiation therapy were excluded from the study (exclusion criteria).
3.2. RNA Extraction and cDNA Synthesis
Tumor and healthy tissues were kept paraffin-embedded; 50 mg of each tissue was transferred to DNAse/RNAse-free sterile microtubes for RNA extraction. RNA was extracted from the collected tissue samples, using a miRNA extraction kit (MN, Germany Cat # 740304). The amount of extracted RNA was measured with a nanodrop device (Thermofisher, US) by calculating the adsorption ratio of 280/260. To evaluate the quality, 200 ng extracted RNAs were run on 0.8% agarose gel and the presence and quality of 18 s and 28 s ribosomal RNA bands were examined. For cDNA synthesis, the Thermo cDNA synthesis kit (Thermofisher, US) was used. All steps were based on the manufacturer's instructions.
3.3. Real-time PCR
Real-time PCR (RT-PCR) reaction was performed by the cyber green method, using Q Rotor-Gene (Qiagen, Germany) device. The β-actin gene (forward: 5'-CACCCAGCACAATGAAGATCAAGAT-3'; reverse: 5'-CCAGTTTTTAAATCCTGAGTCAAGC-3') was considered the internal control gene. The sequence of primers for lncRNA GHET1was forward: 5'-AGTCAGCTCCTACAGGGTG-3' and reverse: 5'-TCCTTAGGTGGTTTCTGTTC-3' and for lncRNA, ZXF2 was forward: 5'-CACCCAGGTCAGAGA-5' and reverse: 5'-TGGAAGGGACACTAGAAGAAGAAT-3'.
The reaction mixture consisted of master mix (7 µL), forward (0.2 µM) and reverse (0.2 µM) primers, 1 µL cDNA, and 6 µL double distilled water. The real-time PCR temperature program for lncRNA cDNA amplification included 95°C for 12 minutes for denaturation and enzyme activation, 95°C for 15 s for denaturation, 60°C for 20 s for annealing primers, and 72°C for 30 s for extension. Eventually, the gene expression was normalized to the β-actin gene, and results were calculated as fold change relative to the control (n = 5 per group and time point) (20). The melting curve was, then, examined.
3.4. Statistical Analysis
One-way analysis of variance (ANOVA) and Tukey post hoc test were used to analyze the data obtained from lncRNA GHET1 and lncRNA ZXF2 expressions. χ2 test was used to investigate the relationship between the expression of lncRNA GHET1 and lncRNA ZXF2 with clinicopathological variables. The KS normality test was performed to assess the data's normality at a significant level of < 0.05. Receiver operating characteristic (ROC) analysis was used to assess the biomarker potential of the genes under consideration.
4. Results
4.1. Clinicopathological Features
The age range of the 30 patients with OSCC varied from 26 to 86 years, with a median of 64.13. Around 73% of patients were men; 56.6% of tumors were in grade 1 and most tumors were in stages II and III (Table 1).
| Attributes | Cases (%) |
|---|---|
| Patient number | 30 |
| Age | |
| Median (range) | 64.13 (36 - 76) |
| Gender | |
| Male | 18 (60) |
| Female | 12 (40) |
| Grade | |
| I | 17 (56.6) |
| II | 13 (43.4) |
| Stage | |
| I | 4 (13.3) |
| II | 10 (33.3) |
| III | 9 (30) |
| IV | 7 (23.3) |
4.2. LncRNA GHET1 and LncRNA ZXF2 Expressions
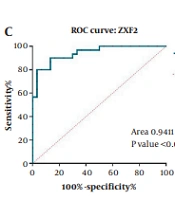
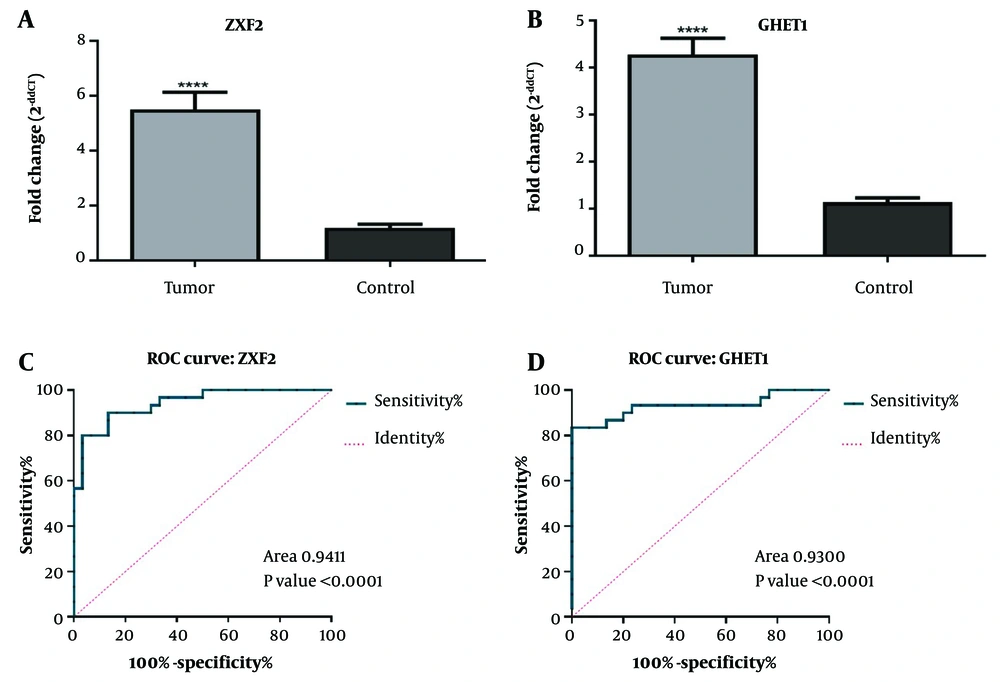
The results of the present study showed that the expressions of both lncRNA GHET1 and lncRNA ZXF2 (Figure 1A and B) in OSCC tumor tissue of patients were higher than in healthy tissue (P < 0.0001). Therefore, it seems that the overexpression of lncRNA GHET1 and lncRNA ZXF2 have regulatory roles in OSCC. Also, the biomarker potential of lncRNA GHET1 and lncRNA ZXF2 for diagnosing OSCC were studied, using the ROC test and the results showed that both lncRNA GHET1 (area under curve 0.94, P < 0.0001) (Figure 1C) and lncRNA ZXF2 (area under the curve 0.93, P < 0.0001) (Figure 2C) have high sensitivity and specificity for OSCC diagnosis.
Comparison of long non-coding RNA (lncRNA) ZXF2 (A); and lncRNA gastric carcinoma highly expressed transcript 1 (GHET1) (B) expressions in oral squamous cell carcinoma (OSCC) tumor and normal tissues. Receiver operating characteristic (ROC) analysis to determine the biomarker potential of lncRNA ZXF2 (C); and lncRNA GHET1 (D) in OSCC diagnosis. **** P-value < 0.0001
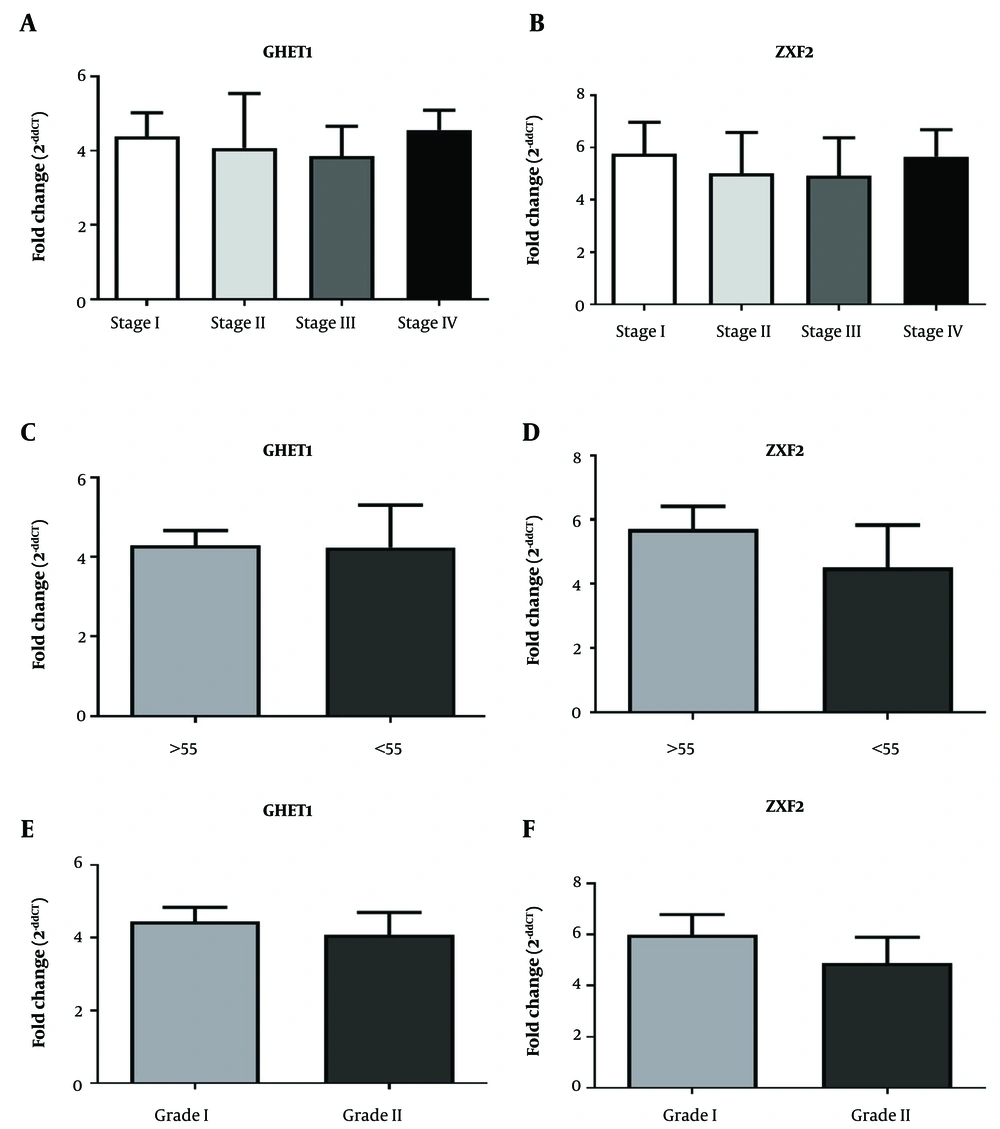
4.2. LncRNA ZXF2 and LncRNA GHET1 Relationships with Clinicopathological Attributes
The study of the relationship between lncRNA GHET1 and lncRNA ZXF2 expressions with clinicopathological variables such as stage, age, and grade in OSCC patients showed that there was no relationship between lncRNA GHET1 and lncRNA ZXF2 expression with these variables (P > 0.05) (Figure 2).
5. Discussion
Oral squamous cell carcinoma is the most common oral cancer and is a serious health problem in most communities (21, 22). Smoking and alcohol abuse are important risk factors for OSCC (23, 24), leading to mutations in genes that regulate cell growth and ultimately lead to severe cell proliferation, abnormal keratinization, abnormal epithelial proliferation, increased cell motility, and angiogenesis (25).
In most cases, OSCC is diagnosed in advanced stages, which significantly reduces the survival rate (26). Therefore, it is necessary to identify the disease in the early stages by introducing specialized biomarkers to properly control prognosis and reduce mortality (27, 28). Thus, in recent years, researchers have evaluated the biomarker potential of lncRNAs in a variety of cancers. The biomarker potential of lncRNA GHET1 and lncRNA ZXF2 in OSCC was verified in this investigation. However, no significant relationship was observed between lncRNA GHET1 and lncRNA ZXF2 expression with the age of patients, stage, and grade tumor.
LncRNA ZXF2 is located at 8q24.28, next to the c-Myc gene, and their interaction has been demonstrated in cell cycle progression, proliferation, migration, and tumor cell invasion (19). Knockdown of lncRNA ZXF2 inhibited c-Myc gene expression and, thus, prevented cancer progression (19). Also, the knockdown of lncRNA ZXF2 increased the tumor-blocking gene E-cadherin (19). Nevertheless, overexpression was associated with poor lymph node metastasis and prognosis (19). Thus, lncRNA ZXF2 plays an oncogenic role in a variety of cancers. In the present study, overexpression of lncRNA ZXF2 was observed in OSCC tumors compared to normal tissue, which may indicate its oncogenic role in lncRNA ZXF2.
In the present study, for the first time, lncRNA GHET1 was shown to overexpress in OSCC. The oncogenic role of lncRNA GHET1 in other cancers such as hepatocellular carcinoma (15), colorectal (17), and gastric (18) cancers has been demonstrated. The knockdown of lncRNA GHET1 has led to a reduction in cancer cell proliferation and metastasis in a variety of cancers (15, 16). However, its overexpression has led to metastasis and poor prognosis as well as resistance to chemotherapy (18). Therefore, according to the mentioned results, it can be stated that lncRNA GHET1 has an oncogenic role in OSCC and can be used as a biomarker for OSCC diagnosis.
In a study by Tang et al., The relative abundance of a set of lncRNAs was studied in tissue or saliva samples from OSCC patients. HOTAIR, NEAT-1, and UCA1 were expressed at greater levels in metastasizing tumors than in others, although MEG-3 expression was downregulated. Importantly, there is no relationship between the expression of these lncRNAs and the gender or age of the patients, indicating that lncRNA is a possible independent risk factor and diagnostic biomarker of OSCC (29). Only HOTAIR was found in saliva with a statistically significant difference from the other lncRNAs studied in this investigation, notably in samples from patients with lymph node metastases. Metastasis is the leading cause of death from OSCC (30). However, in the present study, both lncRNA GHET1 and ZXF2 can be suggesting biomarkers for the diagnosis of OSCC. Han et al. showed that by raising the G1 phase rate, lncRNA GHET1 knockdown may limit cell proliferation, invasion, and migration while enhancing cell death. They also looked into how the lncRNA GHET1 influences breast cancer development. Moreover, in gastric cancer, c-Myc is linked to the lncRNA GHET1. In this study, in cancer tissues, clinical data revealed favorable associations between GHET1 and c-Myc. A c-Myc agonist decreased the antitumor effects of GHET1 knockdown in vitro and in vivo. Our findings suggest that the lncRNA GHET1 is linked to c-Myc expression in breast cancer (31). In another study, Song et al. meta-analyzed study, discovered the potential clinical uses of GHET1 for cancer prediction and tumor progression. The findings indicated that the lncRNA GHET1 can be used as a predictive biomarker in Chinese cancer patients (32).
In a study by Binang et al., the results showed that lncRNA ZXF2 is a potential biomarker for the detection of gastric cancer at the molecular level and may be used as a potential target for gastric cancer therapy. Moreover, it may be utilized for prognostic predictions (33).
5.1. Conclusions
In general, it can be concluded that overexpression of lncRNA ZXF2 and lncRNA GHET1 occur in OSCC and these can be utilized for prognostic predictions in OSCC.