1. Background
Malignancies are currently the leading cause of mortality worldwide, imposing a significant burden on human health and the global economy. Several therapeutic approaches, such as surgery, chemotherapy, and radiotherapy are traditionally used for cancer treatment, and these treatments have been found to be effective for some types of malignancies. Despite the success of these treatments, cancer treatment still remains a major clinical challenge and there is a great need for finding novel methods of treatment for different cancers.
Many research efforts have been made to find effective anti-cancer treatments based on immunotherapeutic approaches, such as virotherapy. Today, some viruses from different viral families are receiving increasing attention as oncolytic therapeutic agents (1). In a previous study, the wild-type Urabe strain of mumps virus (MuV) was investigated clinically for its oncolytic activity (2). Since the discovery of the oncolytic activity of MuV in the mid-1970s, different MuV strains (natural/chimeric and wild/attenuated) have been found to be effective against a variety of cancers (3-11). According to previous studies, a combination of MuV and measles virus (MeV) vaccine strains can exert synergistic anti-tumor effects on cancerous cells (12, 13).
MuV is an enveloped virus having a linear negative-sense RNA genome, and belongs to the family paramyxoviridae, and the genus Orthorubulavirus. Until now, several MuV vaccine strains have been developed in different countries (14). The RS-12 strain-based vaccine was developed in Iran from a wild MuV, which was originally isolated from a throat-wash specimen of a patient with typical symptoms of mumps, including fever and bilateral parotitis, without any other complications (15). The virus was isolated in green monkey kidney cells (GMKCs) and underwent several passages at a low temperature in normal human diploid cells (MRC-5), producing an attenuated phenotype. Further analyses showed that this strain belonged to the MuV genotype H (16), and subsequently, the presence of at least two viral subpopulations was identified in this strain (17). The RS-12 strain is adapted to human diploid cells and grows optimally in normal diploid cells, such as MRC-5.
2. Objectives
The oncolytic potency of such viruses (normal cell-adapted strains) may be improved through its adaptation to cancer cells. Therefore, in this study, we aimed to evaluate the oncolytic activity of the MuV RS-12 strain-based vaccine after its adaptation to cancer cells via serial passaging. For this purpose, serial passaging of the RS-12 strain was carried out in cancer cells (HT1080) eight times. A cancer cell-adapted, the high-growth variant was isolated by terminal endpoint serial dilutions. The regions encompassing four known heterogenic positions of the genome were sequenced and analyzed to demonstrate the genetic homogeneity of the variant. Besides, its oncolytic activity was compared with its parental vaccine working seed in vitro. Finally, the apoptosis-inducing effects of the variant on cancer cells were quantified by flow cytometry.
3. Methods
3.1. Virus and Cells
The history of the development of attenuated MuV RS-12 strain has been described previously (15). In the present study, this strain was available in-house. The current stock was propagated in the MRC-5 cell substrate and extracted from the cell debris through a 0.2-µm syringe filter (Sartorius, Goettingen, Germany). The HT1080 cell line was obtained from the Iranian Biological Resource Center (IBRC; Tehran, Iran). The MRC-5 and HeLa cells were also available in-house. The cell stocks were routinely checked for mycoplasma and other bacterial contaminations using standard methods. The cells were then grown and passaged using Dulbecco’s Modified Eagle Medium (DMEM; Sigma-Aldrich, St. Louis, MO, USA), containing 10% fetal bovine serum (FBS; Gibco, Waltham, MA, USA) at 37°C.
3.2. Isolation of Cancer Cell-adapted MuV RS-12 Variant
To adapt the RS-12 strain-based vaccine to cancer cells, it was passaged eight times in the HT1080 cell line at 33°C, with titration in every passage. Next, the cancer cell-adapted variant was isolated via two terminal endpoint dilutions of the virus in the HT1080 cells. The cancer-adapted virus was ten-fold diluted in DMEM serially, and 0.2 mL of each dilution was inoculated for one hour at 33°C in the HT1080 cells. Next, 50 mL of DMEM has added again, and the cells were incubated at the same temperature. The infected cell cultures were monitored daily for any signs of alteration in the microscopic appearance of the cells (i.e., cytopathic effect [CPE]), especially the syncytium and cell degeneration, using an inverted microscope.
The supernatants from each flask were further passaged onto the same confluent cell type under the same conditions and temperature until the emergence of the CPE. The cell cultures were also examined by the classical hemadsorption assay, using 0.4% guinea pig erythrocytes (18). The final dilution of the virus, which indicated the CPE and/or hemadsorption, was harvested as the cancer cell-adapted variant. The variant underwent two additional passages in the HT1080 cells to generate the seed stock. Subsequently, the harvested stock was subjected to quality control (QC) tests, including sterility and titration tests. The virus stock was also used for further analyses, including genetic homogeneity, in vitro cytotoxicity, and apoptotic potency tests.
3.3. Genetic Homogeneity Analysis
To obtain the sequences of the regions encompassing four known heterogenic MuV RS-12 positions, before and after viral adaptation to cancer cells, viral RNA extraction was performed, using the FavorPrep™ Viral Nucleic Acid Extraction Kit (Favorgen Biotech, Taiwan) according to the manufacturer’s instructions. The viral RNAs were then reverse-transcribed into first-strand cDNA, using a random hexamer primer. Reverse transcription (RT) was carried out, using the cDNA Synthesis Kit (YTA, Tehran, Iran) according to the manufacturer’s instructions.
The synthesized cDNA was used as a template in the polymerase chain reaction (PCR) assay, as previously described (17). Briefly, the PCR reactions were performed in the Mastercycler® Thermal Cycler (Eppendorf, Hamburg, Germany), using three primer pairs: Forward primer MuP1452+ (TGGAGGAATCAGATGACG)/reverse primer MuP3274- (GAGGAATTTTGATCTGTG); forward primer MuM-2720+ (GGTGACCCAAATAAAGAATG)/reverse prime MuM-4122- (TGTGACCGCCTGCATGGA); and forward primer MuL-12164+ (GTATCACTTAAATCAGCACTC)/reverse prime MuL-13713- (CTTGGGAGAGAGTATTTC). The reactions were performed in 0.2-mL microtubes containing 1X PCR buffer, 5 µL of cDNA, 1.5 mM MgCl2, 0.5 µM of each oligonucleotide, 1 U Smart Taq DNA polymerase (CinnaGen Co., Tehran, Iran), and nuclease-free water in a final volume of 50 µL.
The cycle program was set at 95°C for five minutes, followed by 30 cycles (95°C for one minute, 46°C for one minute, and 72°C for two minutes) and a final extension step at 72°C for 15 minutes. The PCR amplicons were subjected to direct sequencing after purification on Agarose gel, using the FavorPrep GEL/PCR Purification Mini Kit (Favorgen Biotech, Taiwan). To verify the presence of heterogeneous sites, amplification was performed in triplicate for each virus and the PCR amplicons were sequenced separately. All sequencing reactions were performed by Bioneer Corporation (Daejeon, Korea).
3.4. In Vitro Cytotoxicity
The MRC-5, HT1080, and HeLa cells (5000 cells/well) were seeded in 100 µL of DMEM (Sigma-Aldrich, St. Louis, MO, USA), supplemented with 5% FBS (Gibco, Waltham, MA, USA), and incubated for 24 hours at 37°C in 5% CO2. After 24 hours, the medium was removed and the cells were infected with viruses at different multiplicities of infection (MOI = 0, 0.002, 0.02, 0.2, and 2). After 96 hours, the medium was removed and replaced with a fresh medium. Next, 10 µL of an active 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) solution (2.5 mg of MTT and 1 mL of PBS) was added to each well, and the plates were incubated for four hours at 37°C in 5% CO2. The medium was then replaced with 100 µL of dimethyl sulfoxide (DMSO). After one hour, the optical density (OD) was recorded on an ELISA plate reader (Dynex MRX II, Chantilly, VA, USA) at 570 nm (630 nm as reference). The results were expressed as the relative viability of the cells, compared to untreated control cells. All data were presented as mean ± SD of three separate experiments, with six wells in each experiment.
3.5. Apoptosis Measurements
Apoptosis and cell viability were measured by annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) double staining. The measurements were performed on day four post-infection, using the FITC-Annexin V Apoptosis Detection Kit with PI (BioLegend, San Diego, CA, USA), according to the manufacturer’s instructions. Annexin V-FITC and PI were used to distinguish viable cell populations (Annexin V-, PI-), early-phase apoptotic cells (annexin V+, PI), late-phase apoptotic cells (annexin V+, PI+), and necrotic cells (annexin V-, PI+). All flow cytometric measurements were carried out at the IBRC (Tehran, Iran). Besides, the cell fluorescence was analyzed using the Flowing Software V.2.5.1. Differences between the median fluorescence intensities were compared using paired t-test, and the level of statistical significance was set at P < 0.05.
4. Results
4.1. Virus Adaptation and Genetic Homogeneity
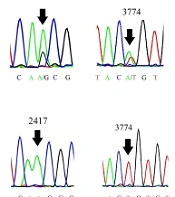
To obtain a virus that grows efficiently in cancer cells, serial passages of MuV RS-12 strain-based vaccine were performed in HT1080 cells. The titer of the strain increased throughout passages in cancer cells, reaching a maximum virus titer of 107.25 cell culture infective dose (CCID50)/mL (Figure 1A). Next, the cancer cell-adapted variant was extracted via terminal endpoint dilutions of the supernatant after the eighth passage in HT1080 cells. To demonstrate the genetic homogeneity, direct sequences of the cancer cell-adapted variant and its parental virus were determined and compared. According to the chromatograms, the RS-12 vaccine working seed exhibited secondary nucleotide peaks at positions 1591, 2417, 3774, and 12977, while cancer cell-adapted RS-12 variant did not show any secondary peaks in these positions (Figure 1B). In these positions, the working seed exhibited major cytosine, adenine, adenine and guanine peaks, respectively, as well as secondary peaks of adenine, guanine, thymine and adenine, respectively, suggesting the presence of at least two viral subpopulations.
(A), The viral yields (the mean of three independent experiments) in different passages (up to the eighth passage) in the HT1080 cell line. (B), The schematic representation of the MuV genome and the four known heterogenic positions of the MuV RS-12 genome. The chromatograms show secondary nucleotide peaks in N, P, M, and L gene sequences of the original working seed. The nucleotide peaks show predominant and minor viral subpopulations (arrows). The analysis of N, P, M, and L gene sequences for the cancer cell-adapted variant indicates a single peak in these positions (arrows).
4.2. In Vitro Cytotoxicity
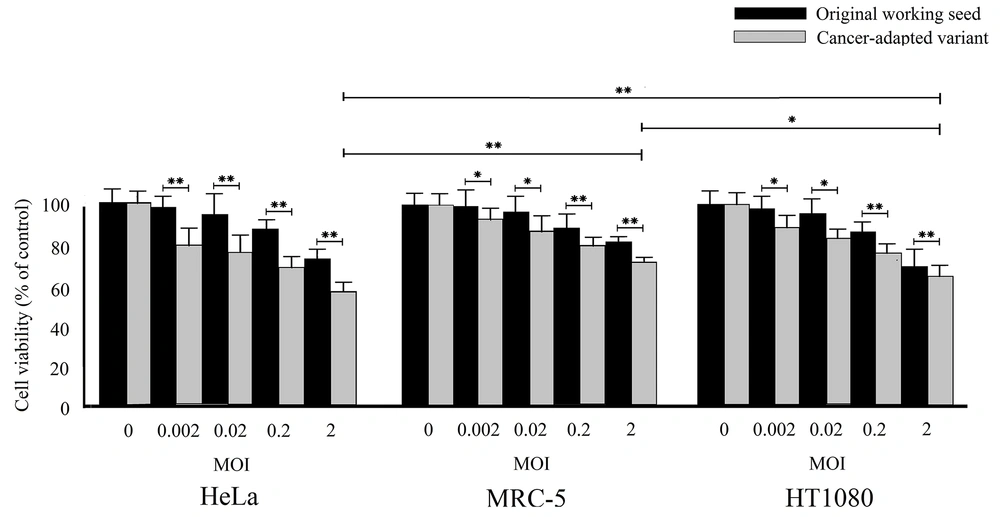
The cytotoxic effects of cancer cell-adapted RS-12 variant and its parental virus working seed were assessed using the MTT assay. Figure 2 presents the MTT assay results for the MRC-5, HeLa, and HT1080 cells after infection with viruses at MOI of 0 to 2. The results showed that both viruses could significantly induce cytotoxicity in these two cancer cell lines. However, the growth of infected cancer cells was significantly inhibited to different degrees. The highest cytotoxicity by the cancer cell-adapted variant was observed in the HeLa cells. These results clearly revealed that the cytotoxicity of the cancer cell-adapted variant was significantly higher than the parental working seed (P < 0.05), even at the lowest MOI (0.002). Overall, the results showed that the HeLa cell line is highly sensitive to cancer cell-adapted RS-12 strain.
The oncolytic effects of the original working seed and the cancer cell-adapted MuV RS-12 variant on the cell viability according to the MTT assay. The cell viability was examined in different cultured cells, including MRC-5, HeLa, and HT1080 cells. The results are shown as mean ± SD of three independent experiments performed in triplicate (*significant difference at P < 0.05; **significant difference at P < 0.001).
4.3. Apoptosis Measurements
Annexin V and PI double staining were performed to distinguish viable cells, early-phase apoptotic cells, late-phase apoptotic cells, and necrotic cells (Figure 3A). As shown in Figure 3B, infection with the RS-12 strain induced a higher percentage of annexin V+/PI- (early-phase apoptotic cells) and annexin V+/PI+ (late-phase apoptotic cells) in the infected HT1080 and HeLa cell lines, compared to the untreated control cells (P < 0.05). The cancer cell-adapted variant showed a higher percentage of apoptotic cells compared to the parental working seed; however, there was no significant difference between the two groups (P > 0.05).
(A) The FSC/SSC plots showing the HeLa and HT1080 cell populations. The representative dot plots of the distribution of necrotic (annexin V-, PI+), viable (annexin V-, PI+), late-phase apoptotic (annexin V+, PI+), and early-phase apoptotic (annexin V+, PI-) cells are shown in control and MuV RS-12 inoculations (one of the three experiments is shown here). (B) The percentages of necrotic, viable, late-phase apoptotic, and early-phase apoptotic cells were calculated in the total cell count. The results are expressed as mean ± SD of three independent experiments performed in triplicate (*significant difference at P < 0.05; **significant difference at P < 0.001).
5. Discussion
In this study, for adaptation of the MuV RS-12 strain-based vaccine to cancer cells, serial passaging was performed in the HT1080 cell line eight times. The results showed an increase in the growth potential of the cancer cell-adapted variant compared to the parental virus. The assessment of growth efficiency in cancer cells revealed that the produced progeny virus titers were approximately one log higher than the titers in normal human diploid cells (MRC-5). Besides, the ability of this variant to optimally replicate at 33°C revealed the maintenance of its temperature sensitivity.
The oncolytic activity of the cancer cell-adapted virus was evaluated in two different cancer cell lines, including the HT1080 fibrosarcoma and HeLa adenocarcinoma cell lines. According to the in vitro analysis, these cell lines exhibited differential sensitivity to the cancer cell-adapted MuV RS-12. The inoculation of the cells at four MOIs showed that the cancer cells were efficiently destroyed by this variant, even at the lowest MOI (0.002). Overall, the hierarchy of cell sensitivity was as follows: HeLa > HT1080 > MRC-5. The HeLa cell line was the most sensitive, whereas the normal human diploid MRC-5 cells were the least sensitive, which might be due to the higher expression of sialic acid (a known MuV receptor) on the surface of cancer cells, compared to the normal diploid cells (19).
Generally, apoptosis is a programmed cellular process, which can be triggered by many factors, such as viral infection (20). The present results showed that the RS-12 variant could efficiently induce apoptosis in two types of cancer cell lines. Additionally, the flow cytometry results revealed that the MuV RS-12 strain not only triggered apoptosis in the cancer cell lines but also induced necrosis. In other studies, the induction of apoptosis and necrosis in different cells has been reported in other MuV strains (3, 11-13, 21). Conversely, some previous studies found that the MuV small hydrophobic (SH) protein could prevent apoptosis by inhibiting the tumor necrosis factor-alpha (TNF-α) signaling (22-24).
Moreover, a previous study showed that the SH protein of MuV had anti-apoptotic activities due to its association with the proteasomal degradation machinery (25). Another study revealed that the V protein of MuV had the potential to modulate apoptosis (26). Besides, MuV can inhibit apoptosis through interferon-alpha (INF-α) and/or interferon-gamma (INFγ) signaling pathways (27-30). On the other hand, the present results indicated that the MuV RS-12 strain is a strong apoptosis inducer in two human cancer cell lines, although its genome contains complete SH and V open reading frames (ORFs), as well as their intergenic regions, and potentially encodes functional SH and V proteins.
The MuV vaccine strains are generally a mixture of genetic mutants in the same strain. Genetic heterogeneity has been observed in different MuV strain-based vaccines (17, 31-36) due to several factors, especially the absence of a proofreading mechanism by the RNA-dependent RNA polymerase (RdRP) during viral genome replication. According to Figure 1B, the chromatogram showed that the parental vaccine seed exhibited two peaks at nucleotide positions 1591, 2417, 3774, and 12977, indicating the presence of at least two different viral subpopulations. On the contrary, the sequences found in the cancer cell-adapted RS-12 variant, isolated by serial dilution, only exhibited subsidiary nucleotide peaks at the same positions (sequences of the minor viral subpopulation), with the exception of position 2417, which represented the major nucleotide peak. However, in the present study, terminal endpoint serial dilutions were used for isolating a discrete variant from mixed viral populations to ensure the homogeneity of the virus stock. The results showed that this method may be a simple and efficient tool to enhance viral homogeneity. Besides, dilute passaging has been occasionally used for selecting the viral variants of different viruses, such as MuV (33).
The obtained findings suggested that different viral variants, whose genomic sequences are closely related but have several different nucleotides, may exhibit different oncolytic potencies. Also, the viral genetic homogeneity may have remarkable effects on the oncolytic potential in vitro. Serial passaging of the virus in cancer cells can increase the viral oncolytic efficacy through genetic modifications that allow for a better replication in these cells; therefore, these modifications may improve the oncolytic potency. One of the heterogenic positions of the original working seed (A2417G) is located in a region around the non-templated nucleotide residues insertion site of the phosphoprotein (P) gene. It has been previously shown that such mutations are associated with alterations in the transcriptional editing accuracy of the P gene, and subsequently, the accuracy of its product expression (37). Another study revealed that the over-attenuated MuV phenotype might be associated with the impaired expression of P protein (38). Overall, it seems that the over-attenuated MuV does not exhibit potent oncolytic or immunotherapeutic activities (4).
5.1. Limitations
There were some limitations to this study. The genetic stability of the developed variant could not be measured in this preliminary study, while the genome stability may be important in maintaining the increased oncolytic potential. Therefore, further comprehensive experimentations, including in vivo analyses, are needed to confirm our in vitro observations regarding the high oncolytic activity of this cancer cell-adapted virus.
5.2. Conclusions
In conclusion, the present study provided preliminary findings regarding the potent oncolytic effects of the cancer cell-adapted MuV RS-12 variant. The results revealed that adaptation of the virus to cancer cells significantly improved its oncolytic potency. Considering the potent oncolytic activity of this variant at a very low MOI, its use for anti-cancer therapy does not seem to require a different formulation in comparison to the routine MuV vaccines; however, further preclinical studies are needed to confirm this finding. Also, a better understanding of the properties of this variant may help us develop a safe, efficient, and cost-effective oncolytic agent with high oncolytic potency.