1. Background
Liver disease is one of the major global health problems and its prevalence is increasing worldwide (1). Hepatocellular carcinoma (HCC) is the most prevalent liver cancer type with nearly 841,000 new cases and 782,000 deaths annually (2). The most important causes of HCC are viral factors including hepatitis B virus (HBV) and hepatitis C virus (HCV), as well as alcohol and fatty liver diseases that vary in different geographical regions. However, HBV has been introduced as the leading cause of HCC, globally (1, 3). Chronic HBV infection has significant roles in HCC promotion through either direct and/or indirect mechanisms (4) including HBV genomic mutations, HBV DNA integration, dysregulation of miRNA, and involvement in epigenetic modifications (5). Following HCC, intrahepatic cholangiocarcinoma (iCCA) is the second most common liver cancer (6) with approximately 15% accounting of liver cancers (2). iCCA is a malignant tumor that originates from epithelial cells of the bile duct (7) and has a high mortality rate due to late diagnosis and aggressiveness (8). Epidemiological studies have shown that the incidence of this cancer has recently increased in Western countries (9). Complicated and multiple risk factors have been introduced for iCCA; congenital and bile duct abnormalities, parasites, Opisthorchis viverrine, inflammatory disorders, chemicals and radiations, toxins, genetic disorders, gallstone, drinking, diabetes, etc.(10, 11), all could lead to chronic inflammation or cholestasis (11). Similarly, chronic viral hepatitis including HBV has been reported to be involved in the development of iCCA (6).
Occult HBV infection (OBI) is an emerging term in which HBsAg is absent, however, HBV DNA could be detected in the liver tissue, peripheral mononuclear cells (PBMC), or serum (12). OBI can be diagnosed in two forms; seropositive (Anti-HBc and/or anti-HBs can be found) and seronegative (Negative for all HBV seromarkers) (13, 14). Covalently closed circular DNA (cccDNA) is the hallmark of a replicative persistent infection rather than a just latent and/or remnant of HBV particles. Traces of cccDNA in the hepatocytes are correlated with viral load (15). Furthermore, decreased level and/or absence of cccDNA is a cause of OBI. Consequently, HBV transcription and protein synthesis are decreased, and eventually, HBsAg turned to undetectable status (16). Around the world, the epidemiology of OBI is in high variation, mostly is much dependent to the endemicity level and rate of HBV among populations (12). Previously findings of OBI among HCC cases are yet limited, as well, this figure has not been regarded well in iCCA. The OBI has been identified in a significantly high rate of HCC patients (More than 40%), moreover, the incidence of HCC among patients with OBI has been estimated as higher rather than cases without OBI (16).
2. Objectives
Though, most similar studies have been focused on blood samples, therefore, a survey of liver samples can be useful. Therefore, we aimed to perform a molecular investigation of OBI within liver tumor samples.
3. Methods
3.1. Study Design, Population, and Samples
This cross-sectional study was conducted on HCC and iCCA in patients referred to hospitals affiliated with Iran University of Medical Sciences. Medical records of patients were checked; all patients were tested for HBV seromarkers using ELISA assay, and those negative for HBsAg were selected. Liver biopsy specimens, including fresh frozen (FF) and archived formalin-fixed paraffin-embedded (FFPE) tissue samples, were used in this study. FFPE samples were collected from January 2013 to March 2020, and FF samples also were collected from April 2017 to April 2020. Clinical, pathological, and demographic characteristics were obtained from hospital records. This study was conducted according to the Declaration of Helsinki. Ethical consideration was approved by Iran University of Medical Sciences Ethics Committee (Ethic code: IR.IUMS.REC.1398.008).
3.2. Preparation of the Samples
Sectioning of FFPE tissue samples was done using sterile and stainless-steel disposable blades. For each sample, eight sections with 15µ thickness were put into sterile 2 mL microtubes. Paraffin removal was performed by putting the sample in 1 ml of xylene (50°C for one hour), and then xylene was removed using washing and drying with 100% ethanol. Furthermore, FF liver tissue samples (needle biopsies) were cut and next were homogenized with a micro pistol.
3.3. DNA Extraction
Afterward, liver tissue samples were subjected to DNA extraction using Tissue DNA Kit (GeneAll®) according to the manufacturer’s protocol with the addition of in-house extra steps. The concentration and quality of extracted DNA were determined by NanoDrop spectrophotometry. The process of DNA extraction and PCR-fidelity was assessed with a PCR assay for an internal housekeeping gene.
3.4. Detection of Occult Hepatitis B Virus Infection
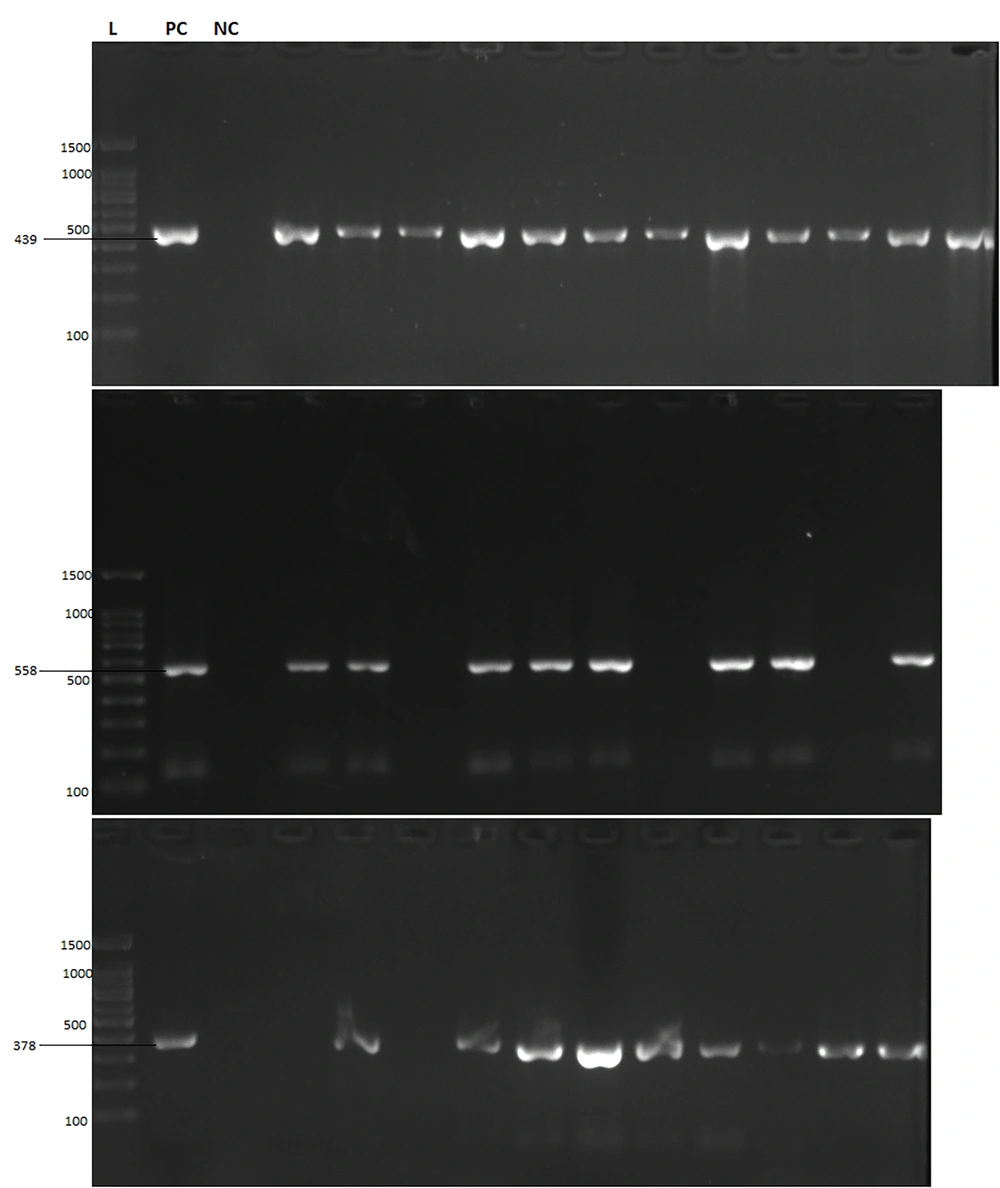
The OBI was detected by performing nested-PCR assay on three HBV genes including S, C, and X, as described previously (17-19). Details of primer sets used are indicated in Table 1. In order to deal with complications of DNA breaks of FFPE samples and optimization of OBI detection, the PCR assays were set in a wide range of PCR amplicons from 225 - 795 base pairs (bp). Briefly, for an outer run of PCR, 25 - 50 ng of extracted DNA was used in a mix of 20 pmole/mL of each external primer, 12.5 μL of PCR mix 2X (Amplicon, red), and sterile water in a final volume of 25µL. Two microliters of the first PCR product were forwarded to nested-PCR in reaction conditions similar to the first run.
| Primer Name | Sequence (5' - 3') | Region/Gene | Position a | Product Size | Ref |
|---|---|---|---|---|---|
| BC1 | HBV Core | 531 | (18) | ||
| F | CTGTTCAAGCCTCCAAGCTG | 1860 - 1879 | |||
| R | GAGGCGAGGGAGTTCTTCTT | 2371 - 2390 | |||
| BC2 | 378 | ||||
| F | TTRCTCTCKTTTTTGCCTTCTG | 1943 - 1964 | |||
| R | TAGGATAGGGGCATTTGGTG | 2301 - 2320 | |||
| HBX1 | HB-X | 795 | (20) | ||
| F | GCTTTCRCYTTCTCGCCAAC | 1087 -1106 | |||
| R | CACAGCTTGGAGGCTTGAAC | 1881 - 1862 | |||
| HBX2 | 558 | ||||
| F | ACTCCTWGCCGCTTGTTTYG | 1278 - 1297 | |||
| R | GGCAGAGGTGAAAAAGTTGC | 1835 - 1816 | |||
| HS1 | HBs | 484 | (17) | ||
| F | AGAACATCGCATCAGGACTC | 159 - 178 | |||
| R | CATAGGTATCTTGCGAAAGC | 642 - 623 | |||
| HS2 | 439 | ||||
| F | AGGACCCCTGCTCGTGTTAC | 181 - 200 | |||
| R | AGATGATGGGATGGGAATAC | 619 - 600 | |||
| BSq | HBs | 225 | (17) | ||
| F | TCGTGGTGGACTTCTCTCAA | 254 - 273 | |||
| R | AGGACAAACGGGCAACATAC | 478 - 459 | |||
| Bcc | cccDNA | 260 | (21) | ||
| F | ACTCTTGGACTCCAGCAATG | ||||
| R | CTTTATACGGGTCAATGTCCA | ||||
| P | FAM- CTTTTTCACCTCTGCCTAATCATCTCWTGTTCA-TAMRA |
a The numbering is based the sequence order of whole HBV genome, accession number ON093165.
3.5. Genotyping and Mutation Analysis
The sequence of HBV S gene was determined using direct Sanger sequencing of PCR products. The sequence data were analyzed using MEGA7, CLC workbench and BioEdit software. Determination of genotype and subgenotype was performed in the dataset of Geno2pheno. Phylogenetic analysis was performed using reference sequences of all known genotypes and subgenotypes of HBV in the benefit of a neighbor-joining method for constructing phylogenetic trees.
3.6. Detection of Viral Load and cccDNA
The HBV DNA load was determined using a real-time PCR assay, as described previously (20). Detection of HBV cccDNA in liver tissue specimens was conducted by a probe-based real-time PCR method, using a Rotor-Gene device (QIAGEN, Germany), as previously explained (21). The RT-PCR method was conducted using a 25 μL total mixture including 10 mole of the primer-probe mix, 5 μL of the DNA extraction product as template and 12.5 μL Master Mix Probe (Yekta Tajhiz Azma, YTA CO, Iran). The thermoscycling conditions of cccDNA assay consisted of an initial step for 3 minutes at 95°C, followed by 45 cycles at 95°C for 15 seconds and 59°C for 50 seconds.
4. Results
4.1. Population-Based Information
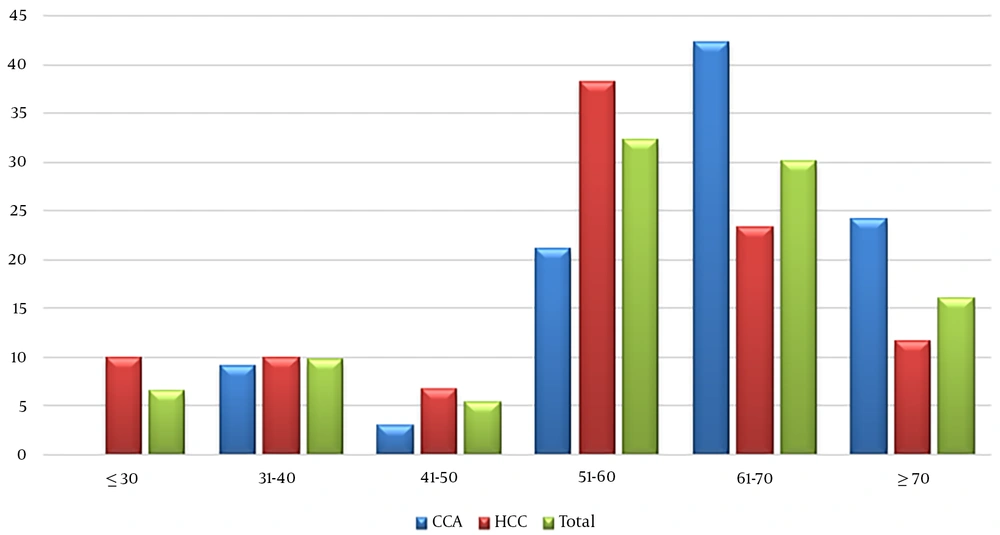
Overall, 93 participants were evaluated, including 60 HCC (Including 48 FF and 12 FFPE) and 33 iCCA (Including 18 FF and 15 FFPE). The mean ± SD age of the studied participants was 57.2 ± 13.8 years, ranging from 22 to 85 years (Figure 1). The mean age in iCCA group was significantly more than HCC (62.1 ± 12.3 and 54.5 ± 13.9 years of old among iCCA and HCC, respectively; P: 0.01). Among the participants, 66.7% were males and the rest were female; this figure among HCC and iCCA groups was 68.3% and 63.6%, respectively. The result of serological records showed that all the patients were negative for HBsAg. The prevalence of anti-HBc among HCC and iCCA were 16.6 and 9.1%, respectively. The status of anti-HBs has been checked among 83.3% and 75.75% of patients with HCC and iCCA, respectively; among which 80% and 72% had anti-HBs titers more than 10 IU/mL (Table 2).
| HCC, No. (%) | CCA, No. (%) | Total, No. (%) | P-Value | |
|---|---|---|---|---|
| Total | 60 (47.6) | 33 (26.2) | 93 | |
| Anti-HBc | 10 (16.7) | 3 (9.1) | 13 (14) | 0.158 |
| Anti-HBs | 40 (66.6) | 19 (57.6) | 59 (63.4) | 0.086 |
| HBV S gene PCR | 11 (18.3) | 3(9.1%) | 14 (11.1) | 0.126 |
| HBV C gene PCR | 6 (10) | 0 | 6 (6.4) | 0.066 |
| HBV X gene PCR | 9 (15) | 3(9.1) | 12 (12.9) | 0.194 |
| OBI | 11 (18.3) | 3 (9.1) | 14 (15) | 0.098 |
4.2. Prevalence of Occult Hepatitis B Virus Infection
First, all samples were checked in terms of PCR-fidelity of extracted DNA and optimization for PCR. There were some FFPE samples that failed in PCR of the internal control gene. Second, the process of extraction was repeated to obtain an efficient yield of DNA. To detect OBI, three independent PCR assays amplifying HBV S, X, and C genes were used (Figure 2). Table 2 shows the detection rate of three HBV genes among the samples. The results of PCR tests demonstrated that among 93 samples, 14 (15%) were positive for OBI. The prevalence of OBI among patients with HCC and iCCA was 11/60 (18.3%) and 3/33 (9.1%), respectively. The rate of OBI was significantly higher among subjects positive for anti-HBc, but had no association with anti-HBs (Table 3).
| HCC, No. (%) | iCCA, No. (%) | Total, No. (%) | P-Value | |
|---|---|---|---|---|
| Observation rate of OBI | ||||
| Anti-HBc | 0.033 | |||
| Positive | 4/10 (40) | 1/3 (33.3) | 5/13 (38.5) | |
| Negative | 7/50 (14) | 2/30 (6.6) | 9/80 (11.2) | |
| Anti-HBs a | 0.142 | |||
| Positive | 8/40 (20) | 1/19 (5.2) | 9/59 (15.2) | |
| Negative | 3/10 (30) | 2/5 (40) | 5/15 (33.3) |
a Percent were given within participants with an anti-HBs test; those with anti-HBs titers more than 10 IU/mL were considered positive for anti-HBs.
4.3. cccDNA and Viral Load
The result of real-time PCR assay was analyzed and showed that the mean viral load of HBV among the OBI cases was 1.3 × 105 ± 1.4 × 102 copies/µL of extracted DNA, ranging from 784 to 1.8 × 106 copies/µL. The molecular test of cccDNA showed that among cases with OBI findings, 5 (35.7%) were positive for cccDNA, and the rest showed negative results.
4.4. Genotyping of HBV
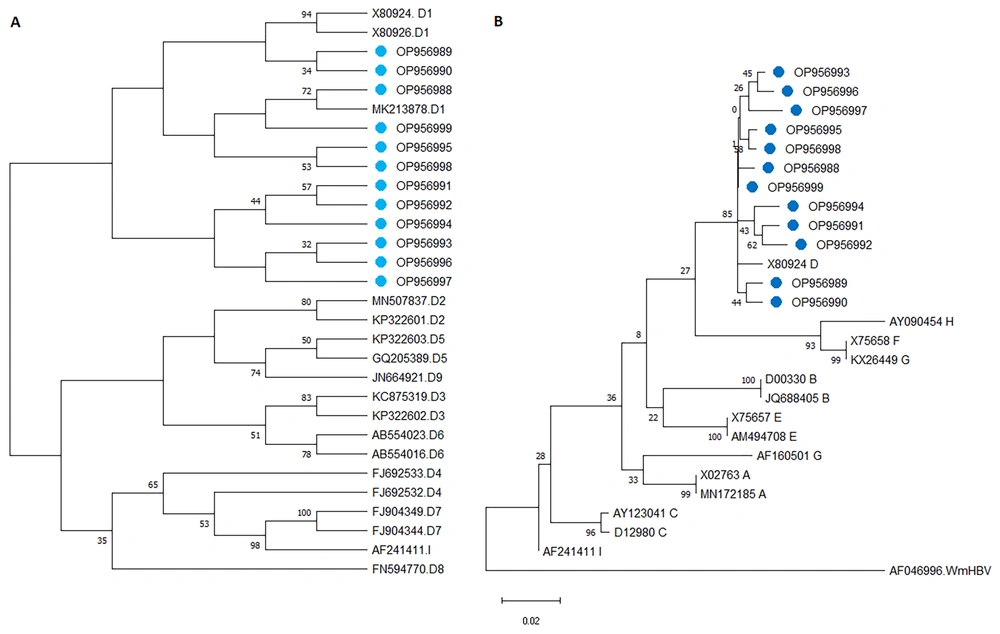
The PCR product of the small S gene was subjected to direct Sanger sequencing. The raw sequence data were observed and trimmed in Chromas and BioEdit software. Next, the sequences were submitted to the gene bank of NCBI (The accession numbers defined for sequences of the current study are OP956988 to OP956999). Primarily, the sequences were searched in the blast tool of NCBI, and then the determination of genotypes and subgenotypes was performed in the dataset of Geno2pheno. The analysis of the nucleotide sequence of the HBV S gene showed that all the isolates of current project belonged to the HBV genotype D, subgenotype D1.
For the construction of a phylogenetic tree in Mega7 software, firstly model selection was done; the “Kimura 2-parameter” model using a discrete Gamma distribution had the lowest Bayesian Information Criterion score was considered to describe the substitution pattern the best. For each model, the AIC value (Akaike Information Criterion, corrected), Maximum Likelihood value, and the number of parameters (including branch lengths) were also obtained (22). The results of phylogenetic analysis are demonstrated in Figure 3.
The phylogenetic tree was constructed with Mega 7 software. The evolutionary history of the HBV S gene was inferred using the neighbor-Joining method. A, Phylogenetic tree for determination of HBV genotypes (cases labeled with a blue circle are isolates of the current study, accession numbers and genotypes are given in front of branches, wooly monkey HBV was used to root the tree). B, Phylogenetic among the subgenotypes of HBV.
4.5. Analysis of Mutations
The results of partial sequencing of the HBsAg coding region were further analyzed and translated to the corresponding amino acid sequence (BioEdit software). In the HBsAg protein, T45N (33.3%) and P105A (25%) were the most prevalent substitutions, furthermore, Q129H, A17P, F20S, P29Q, and C138S mutations were observed in 16.6% of 12 cases (Figure 4). As the S gene is completely overlapped in the RT domain of Pol gene, the counterpart mutations in the RT domain from amino acid 24 - 150 were determined. Most nucleotide substitutions in the ORF of S gene were silent, although they were missense in RT: N53K (33.3%), Y54H (25%), L80I (33.3%), A113G (25%) were the most prevalent missense mutations of RT region (Figure 4).
5. Discussion
This study investigated the OBI in liver biopsy samples among Iranian patients with HCC and iCCA for the first time. There is growing interest on OBI due to its clinical significance. Its most important features include its ability to transmit, reactivation, progression to chronic liver diseases, and development to liver cell carcinoma (12). In patients with chronic HBV infection, the status of HBeAg, HBsAg and viral load are known risk factors for the development and progression to HCC (23, 24). The direct relevance between OBI and HCC is controversial. While, some studies have not been able to explain the direct cause, other studies have reported an association between OBI and HCC (25-27).
In this study, the prevalence of OBI among HCC was 11/60 (18.3%). There were similar studies with higher rates of OBI; in Hong Kong, 24 out of 33 with cryptogenic HCC (72.7%) were found with OBI (28). Another study In China reported it among HCC patients 19.8% (138/698) (29). Although, the higher endemicity level of HBV is noticeable in those regions. Different results are reported by other studies from different regions as well: 15/80 (20.0%), 3/91 (3.3%), and 45/73 (61.6%) in Taiwan, the United States, and Italy, respectively (26, 30, 31). It seems that the probable cause of these differences is due to the diverse rate of HBsAg prevalence in studied regions; as well variable distribution of HBV genotypes should also be considered.
Another finding of the current study was that the prevalence of OBI among iCCA was 3/33 (9.1%). Recently, a significant correlation has been reported between chronic HBV infections and iCCA progression (6). HBV is introduced as a major risk factor for of iCCA in China, Taiwan, and South Korea (32-34). Pollicino et al. have found that 61.7% patients with iCCA were positive for HBV DNA, and cccDNA has been detected in 34.5% of liver samples (6) which is much higher than the figure of the current study. The term OBI in correlation to iCCA is not well studied and needs more investigations globally.
Occult HBV infection appears to have roles in HCC and either iCCA. Furthermore, the presence of cccDNA in the liver is representative of replicative infection that persists in the hepatocytes. This necessitates more precise HBV DNA testing and evokes the requirement for multi-target PCR assays, at least three sites in the HBV genome for detection of OBI (12). However, we performed sensitive nested-PCR assays on three regions of S, X, and C genes to overcome very low replication and/or no replication status of persistent HBV infection.
In terms of intrahepatic viral load, in the present study, HBV viral load was observed in low levels among patients with OBI, which is consistent with other studies (25, 35). Previous research reported the high rate of HBV-DNA integration into the hepatocytes that are associated with the detection of cccDNA (36, 37). In this study, cccDNA was detected among 35% of OBI cases and most of them had viral loads lower than 2000 copies/µL. These findings support the fact that in the OBI status, HBV virion is present in so low replicate form.
Previous studies have been strongly suggestive of a correlation between HCC incidence and HBV genotypes that may be due to variation types and mutants of HBsAg, core and HBx (38). In this study, all the specimens belonged to the genotype D and sub genotype D1. This finding is consistent with the results of other studies conducted in Iran. The most prevalent and dominant genotype in Iran is the genotype D (35, 39-41), as well as among Iran's neighboring countries, including Iraq and Pakistan (42, 43).
In brief, this study demonstrated the detection of OBI in HCC and iCCA in the absence of an overt HBV infection. Further research is required to more accurately define the pattern of OBI in carcinogenesis and distinguish the mechanisms by which it displays pro-oncogenic functions. The Co-existence of in-vitro experiments with this kind of study as well as gene expression analysis could be beneficial for better interpretation and clarification of probable roles of OBI in tumor development.
5.1. Conclusions
Our results indicated that OBI could be a risk factor in exacerbations of liver diseases. The observation of HBV-DNA among HCC and iCCA tumor samples with negative HBsAg figure indicate the risk of HBV persistency and requisite more precise molecular methods in patients’ follow-up. Moreover, the liver samples are very helpful specimens for molecular investigations of OBI as the latency home of the virus.