1. Background
Lung cancers are the most common malignancy and the leading cause of cancer-related death in the world (1). Histologically, they are classified into two types, including non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC), accounting for approximately 85% and 15%, respectively (2). The 5-year survival rate in patients with NSCLC has been reported to be below 15% (3). More than 90% of NSCLC-related deaths are attributed to metastasis and high recurrence rates (4). Angiogenesis and proliferation have also been reported to play an important role in the development of most solid tumors, such as NSCLC (5). The dysregulation of microRNAs (miRNAs) has been demonstrated to be involved in the development of tumors, cell proliferation, drug resistance, invasion, and malignancy in lung cancer (6-8). MicroRNAs also play a role in the occurrence of lung cancer by targeting both tumor suppressor genes and oncogenes (7). Among them, microRNA-128, miR-335-5p, and miR-1254 are mainly reported to function as lung cancer cell behavior. Experimentally, microRNA-128 plays an important role in inhibiting human NSCLC tumorigenesis and angiogenesis by directly targeting VEGF-C (9). In addition, miR-128 has been shown to down-regulate in lung cancer tissues, and it induces apoptosis in lung cancer cell lines (10). The miR-335-5p acts as a tumor suppressor, inhibiting migration and invasion. It has been mainly shown to inhibit TGF-β1-induced epithelial-mesenchymal transition in NSCLC (11). In gastric cancer, miR-1254 inhibits cell proliferation, migration, and invasion (12) and exerts oncogenic effects in human breast cancer, as well (12). It has also been reported that miR-1254 attenuates NSCLC growth by suppressing HO-1 expression (13). K-RAS is one of the most notable driver genes in NSCLC, while VEGF signaling plays a critical role in tumor angiogenesis (14). K-RAS mutations occur in approximately 15% to 25% of NSCLCs (15). It has been reported that activation of K-RAS signaling pathways is involved in cell survival and proliferation (16). However, few studies have focused on a promising insight into the role of miRNAs in the molecular biology of K-RAS-driven NSCLC (17). In this case-control study, the important point is that the expression of these 3 markers (miRNA-128, miR-335-5p, and miR-1254), as well as 2 markers (VEGF and K-RAS), have been examined together and simultaneously and can be presented as a diagnostic package, which has not been done so far. On the other hand, no such study has been done with these markers in the native Iranian race. Therefore, it was necessary to carry out this research according to the geography of the region.
2. Objectives
The purpose of this study was to investigate the expression of markers in tissue and blood separately as diagnostic markers, and it is obvious that many factors can play a role in the development of cancer in different ways.
3. Methods
3.1. Ethics Statement
Ethical approval for this study was obtained from the Research Ethics Committee of Shahid Beheshti University of Medical Sciences, Tehran, Iran (IR.SBMU.NRITLD.REC.1399.218).
3.2. Bioinformatics Prediction of MicroRNAs
A systematic research was conducted in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/GEO) to identify suitable miRNA expression profiles in microarray data (18). In the initial step, a search was conducted, using the keywords 'non-small cell lung cancer' and 'serum', yielding 1038 relevant data entries. The obtained data were, then, refined to 1032 entries by applying the “Homo sapiens filter”. Subsequently, by incorporating the “non-coding RNA profiling by array” keyword into the study type, 23 relevant datasets were obtained. Finally, the sample sizes were balanced, and the data were thoroughly evaluated. In addition, miRNA articles related to NSCLC were also considered to find the miRNAs with biomarker potential for this cancer. MicroRNA-128, miR-335, and miR-1254 were selected and their genomic and conserved regions were investigated, using the UCSC Genome Browser to find the peptide precursors that exist in encoding proteins gene. The candidate miRNAs were selected for laboratory confirmation and evaluated for expression and function. The functions of miRNAs in metastasis, proliferation, apoptosis, and angiogenic were confirmed. To evaluate the role of miRNAs related to NSCLC, alterations in miRNA expression were investigated, using the OncomiR database (http://www.oncomir.org).
A total of 11 online predicted databases of target genes at miRWalk 2.0 were used to select suitable target genes for all 3 chosen miRNAs. The genes defined in the aforementioned databases were considered if they were common in at least 2 miRNAs and also found in TargetScan (http://www.targetscan.org/vert_72). Genes with higher scores were ultimately selected as gene targets and assessed, using DIANA TOOLS- mirPath v.3 (http://snf -515788.vm.okeanos.grnet.gr). At the DIANA database (http://diana.cslab.ece.ntua.gr), data from TarBase 5.0 results and data, comprising over 1 300 proven experimentally targets, were utilized.
3.3. Blood and Tissue Sampling
A total of 50 NSCLC patients and 50 healthy subjects, aged between 22 and 70 years, were included in this study. Patients with any chronic and acute inflammatory diseases were excluded. Since genes have different expressions in different tissues, in the present study, the expression of markers in cancerous tissue has been compared with normal tissue, and also the expression of markers in the blood of people with cancer has been compared with the peripheral blood of healthy people. All participants signed informed consent forms. The blood was collected from healthy donors and the NSCLS patients. The collected sample from each patient was divided into two parts and placed in a tube containing EDTA for measuring the serum level of VEGF, or a tube without EDTA for RNA extraction. Serum level of VEGF was measured, using the commercial enzyme-linked immunosorbent assay (ELISA) kit (Bioassay Technology Laboratory, China) according to the manufacturer’s instructions. After surgery, the cancerous tissues were collected from the margin and central tumors of each patient and immediately transferred to a container containing the liquid nitrogen, and stored until RNA extraction.
3.4. RNA Extraction, cDNA Synthesis, and Real time-PCR
Total RNAs were extracted from blood and tissue samples, using RNeasy Midi Kit (QiagenCat. No: 75144) and Cinna pure (Cat No. PR891620) following the respective manufacturer’s protocols. The concentration and purity of RNA were assessed at 260/280 nm absorbance, and isolated RNA with a 260/280 ratio of ~2 was utilized for further experiments. Isolated RNAs (1µg) were reverse-transcribed to cDNA, using aViva 2-step RT-PCR (Cat no.RTPL12) cDNA Synthesis Kit according to the manufacturer’s protocol. This process was performed in triplicate and 3 vials of each cDNA were synthesized.
Specific primers for VEGF, K-RAS, and 18s rRNA were designed, using AlleleID7 software. The primer sequences are provided in Table 1. The mRNA expression levels of K-RAS and VEGF were determined by Real-time PCR, using Cinna GreenqPCR Mix,2X (Cat No.MM2041) containing Evagreen. Since 18s rRNA has been reported to be a stable housekeeping gene, it was used as the internal comparator in parallel with the control sample (19). cDNA synthesis of miRNAs was performed, using the Reverse Transcription System Kit (Zist Royesh, Iran). The expression levels of the dysregulated miRNAs-128, miR-335-5p, and miR-1254 in the lung tissues of the patients with NSCLC were measured, using ABI 7300 Real-time PCR machine (Applied Biosystem, USA) (20, 21).
| Parameters | VEGF-mRNA | K-RAS-mRNA | 18s rRNA |
|---|---|---|---|
| F primer | TGCAGATTATGCGGATCAAACC | CTGCCCAATCCATTAGCCAC | GTAACCCGTTGAACCCCATT |
| Primer length | 22 | 20 | 20 |
| R primer | TGCATTCACATTTGTTGTGCTGTAG | CACAGAGACCAAACCCCTTCT | CCATCCAATCGGTAGTAGCG |
| Primer length | 25 | 21 | 20 |
| Amplicon length | 81 | 700 | 152 |
| Optimal annealing temperature | 59℃ | 59℃ | 53.5℃ |
3.5. Statistical Analysis
SPSS Software (version 22) was used to analyze the data and the data are expressed as mean ± SD. The difference in mean age between two groups was compared by t student-test. Two-sample binomial analysis test was used to find a statistical difference between the two groups for the positive existence of micro-RNAs and mRNA expression levels of K-RAS and VEGF genes in tissue and blood samples, as well as expression of VEGF protein.
4. Results
The mean age of healthy subjects and NSCLC patients was 42.22 ± 8.31 and 46.58 ± 8.32. There was no significant difference between both groups in terms of age.
4.1. MicroRNA-128in Peripheral Blood and Tissues
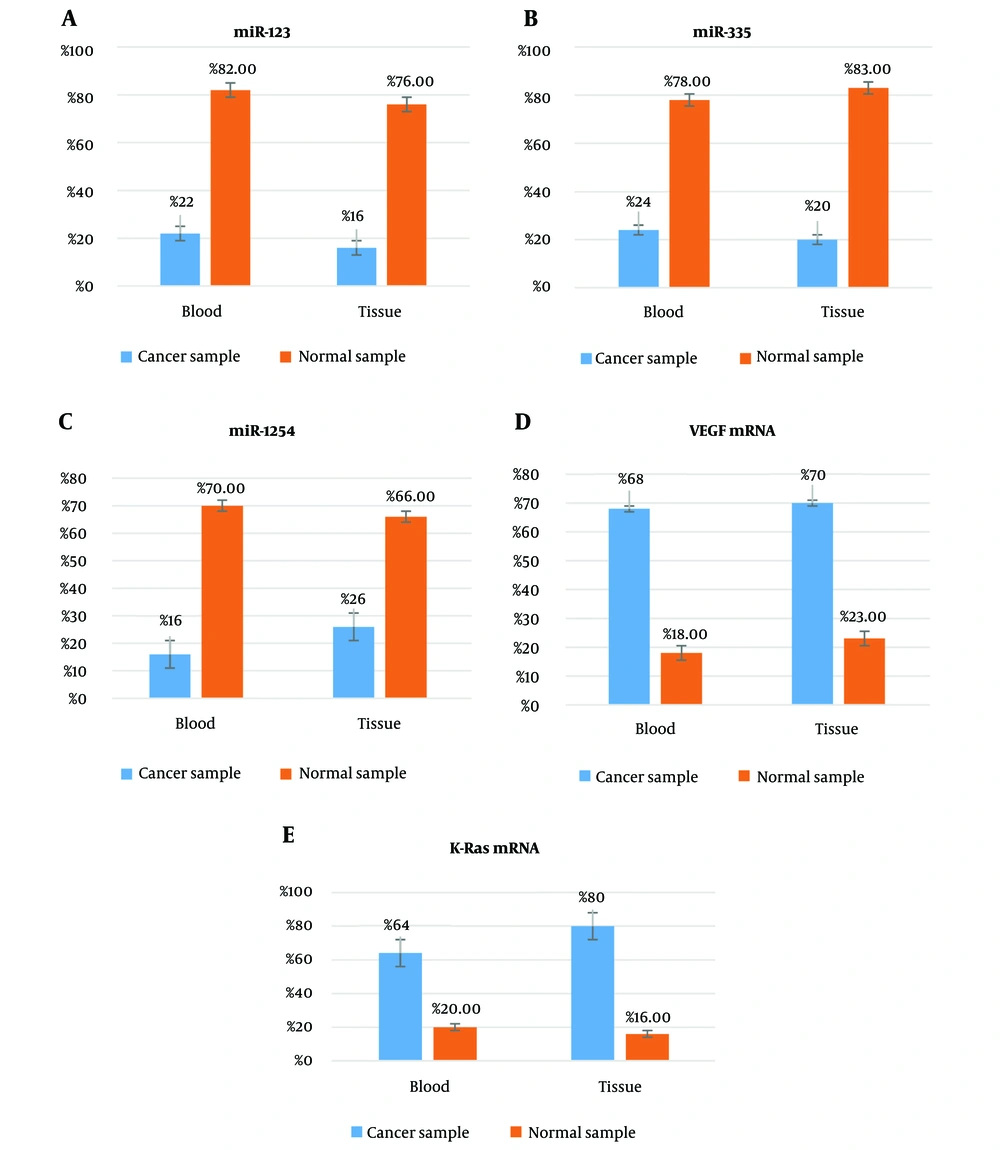
Among the 50 NSCLC patients and 50 healthy subjects, miR-128 was expressed in the peripheral blood of 11 patients (22%) and 41 healthy subjects (82%). There was a significant difference between both groups regarding miR-128 expression in peripheral blood as determined by a two-sample binomial analysis test (P < 0.001). miR-128 expression was observed in 5 cancerous tissue samples (out of 30 samples) (16.66%), as well as in 23 normal tissue samples (out of 30 samples) (76.66%). A two-sample binomial analysis test revealed a significant difference between the two groups regarding miR-128 expression in tissue samples (P < 0.001) (Figure 1A).
4.2. miR-335 in Peripheral Blood and Tissues
We have found that miR-335 is in the peripheral blood of 12 out of 50 patients (24%) and 39 out of 50 healthy subjects (78%). Two-sample binomial analysis test showed a significant difference in the presence of miR-335 in the peripheral blood of healthy subjects compared to NSCLC patients (P < 0.001). The expression of miR-335 was seen in 6 cancerous tissue samples (of 30 samples) (20%), as well as 25 normal tissue samples (of 30 samples) (83.33%). Two-sample binomial analysis test showed a significant difference between both groups concerning miR-335 expression in tissue samples (P < 0.001) (Figure 1B).
4.3. Detection of MicroRNA-1254 in Peripheral Blood and Tissues
The miR-1254 was detected in the peripheral blood of 8 out of 50 patients (16%) and 35 out of 50 healthy subjects (70%). There was a significant difference between the two groups regarding miR-1254 expression in peripheral blood, as determined by a two-sample binomial analysis test (P < 0.001). The expression of miR-1254 was seen in 8 cancerous tissue samples (out of 30 samples) (26.66%), as well as 20 normal tissue samples (out of 30 samples) (66.66%). Two-sample binomial analysis test showed a significant difference between both groups concerning miR-1254 expression in tissue samples (P < 0.001) (Figure 1C).
4.4. VEGF mRNA in Peripheral Blood and Tissues
VEGF biomarker was positive in the peripheral blood of 34 out of 50 patients (68%) and 9 out of 50 healthy subjects (18%). There was a significant difference between the two groups regarding VEGF mRNA expression in peripheral blood, as determined by a two-sample binomial analysis test (P < 0.001). The VEGF mRNA was positive in 21 cancerous tissue samples (out of 30 samples) (70%), as well as 7 normal tissue samples (out of 30 samples) (23.33%). Two-sample binomial analysis test showed a significant difference between both groups in terms of VEGF expression in tissue samples (P < 0.001) (Figure 1D).
4.5. K-RAS mRNA in Peripheral Blood and Tissues
K-RAS biomarker was positive in the peripheral blood of 32 out of 50 patients (64%) and 10 out of 50 healthy subjects (20%). There was a significant difference between the two groups regarding K-RAS mRNA expression in peripheral blood, as determined by a two-sample binomial analysis test (P < 0.001). K-RAS mRNA expression was positive in 24 cancerous tissue samples (out of 30 samples) (80%), as well as 5 normal tissue samples (out of 30 samples) (16.66%). Two-sample binomial analysis test showed a significant difference between the two groups in terms of K-RAS expression in tissue samples (P < 0.001) (Figure 1E).
4.6. The Frequency of VEGF Protein in Normal and Cancer Subjects
VEGF protein was detected in the peripheral blood of 58.19% of the patients (32 out of 50) and 23.3% of the healthy subjects (9 out of 50). Two-sample binomial analysis test showed a significant difference between the two groups in terms of VEGF protein existence in peripheral blood (P < 0.001).
4.7. Comparison of the Expression Levels of MicroRNA-128, MicroRNA-335, and MicroRNA-1254
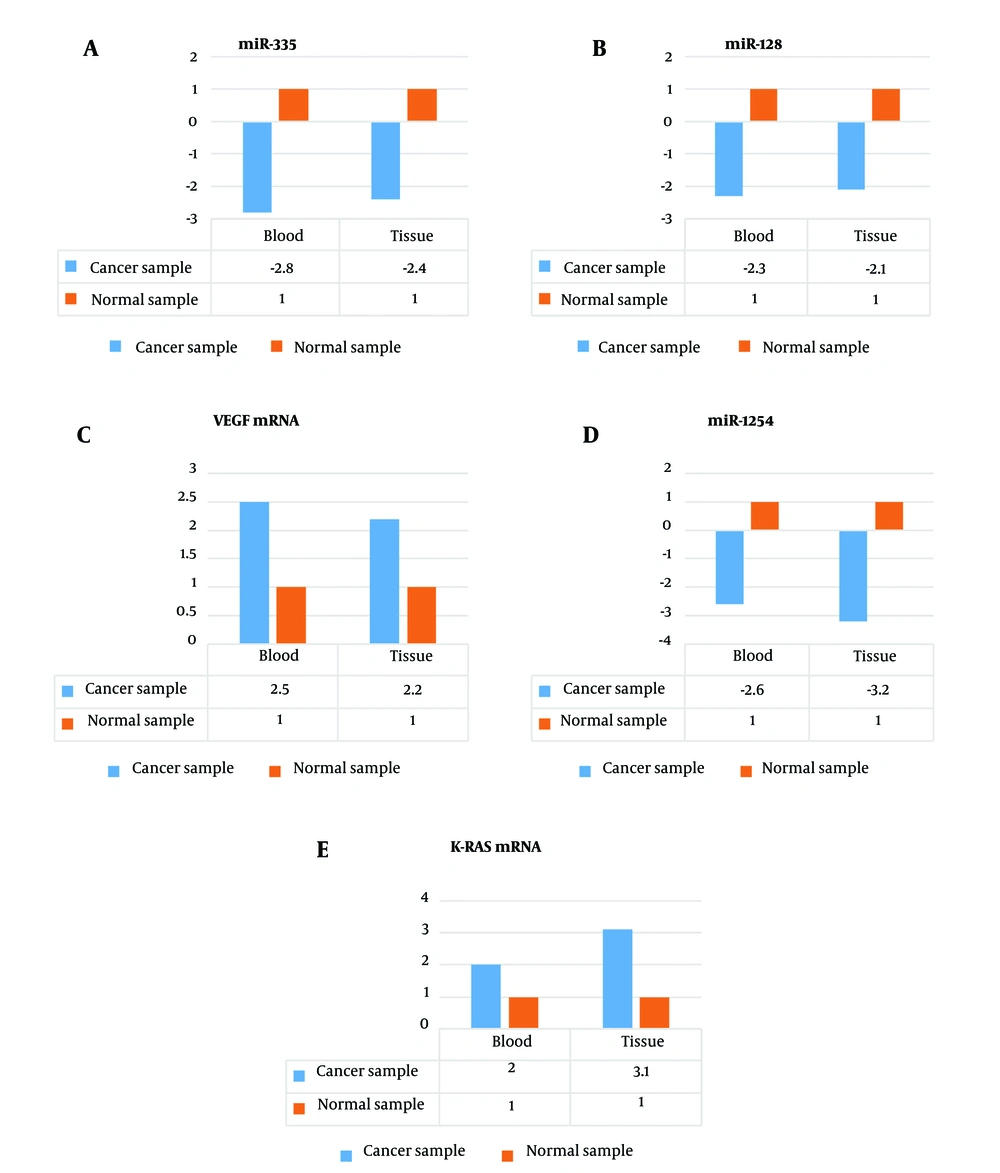
In healthy subjects, the expression of miR-128 in peripheral blood and tissue samples was 2.3 and 2.1 times higher than that in NSCLC patients, respectively (Figure 2A). Similarly, the expression of miR-335 in peripheral blood and tissue samples of healthy subjects was 2.8 and 2.4 times higher than that in NSCLC patients, respectively (Figure 2B). Additionally, the expression of miR-1254 in peripheral blood and tissue samples of healthy subjects was 2.6 and 3.2 times higher than that in NSCLC patients, respectively (Figure 2C).
4.8. Comparison of the mRNA expression levels of VEGF and K-RAS
The mRNA expression level of VEGF in the peripheral blood and tissue samples of the NSCLC patients was 2.5 and 2.2 times higher than that of the healthy subjects, respectively (Figure 2D). On the other hand, the mRNA expression level of K-RAS in the peripheral blood and tissue samples of the NSCLC patients was 2.0 and 3.1 times higher than that of the healthy subjects, respectively (Figure 2E).
5. Discussion
In the present study, the existence and expression levels of miRNA-128, miR-335-5p, and miR-1254 in both peripheral blood and cancerous tissue of NSCLC patients were compared to those in healthy subjects. Additionally, the mRNA and protein expression levels of VEGF (as an angiogenic factor) and K-RAS (as a metastatic and proliferative factor) in both peripheral blood and cancerous tissue of NSCLC patients and healthy subjects were evaluated. It has been estimated that 1.8 million new lung cancer cases, resulting in 1.6 million deaths, occur annually (22) and NSCLC comprises more than 80% of all lung malignancy cases (23).
One of the main findings of the present study was the lower mRNA expression levels of miRNA-128, miR-335-5p, and miR-1254 in the peripheral blood and tissue samples of the NSCLC patients compared to those of healthy subjects. Additionally, these 3 miRNAs were detected in fewer people with NSCLC compared to those of healthy subjects. These findings show that miRNA-128, miR-335-5p, and miR-1254 are down-regulated and decreased in the NSCLC patients compared to healthy subjects.
MicroRNA-335-5p has been shown to be down-regulated in various tumors, including renal, gastric, hepatocellular, and pancreatic cancers (24-27). MicroRNA-335-5p has also been reported to inhibit the metastasis of gastric cancer (28) and cell proliferation, migration, and invasion in colorectal cancer (29). It has been reported that miR-335-5p inhibits cell proliferation and migration of NSCLC, indicating its potential function in NSCLC, particularly in metastasis (30).
MicroRNA-1254 was another studied miRNA in our research, which was down-regulated in the serum and tissue samples. Previous reports have indicated that miR-1254 expression is dysregulated in NSCLC patient serum (31, 32). MicroRNA-1254 has also been shown to induce cell apoptosis and cell cycle arrest, subsequently suppressing NSCLC cell growth (13). The down-regulation of miR-1254 in our patients suggests its involvement in the progression of NSCLC.
The higher mRNA expression level of K-RAS in the peripheral blood and tissue samples of the NSCLC patients compared to that of the healthy subjects was another main finding of the present study. In addition, both K-RAS protein and mRNA were detected in more people with NSCLC than in healthy subjects. These findings suggest that K-RAS can be considered a serum and tissue biomarker in NSCLC patients. K-RAS, an intracellular guanine nucleotide-binding protein or G-protein, belongs to the family of small GTPases, controlling multiple cellular functions such as proliferation, motility or survival, and apoptosis (33). K-RAS mutations are not only the most frequent oncogene aberrations in patients with lung cancers, but the role of the K-RAS oncogene in NSCLC remains unknown (34). It has been previously reported that K-RAS mutations are strictly associated with specific clinical features, such as clinical background, pathological characteristics, and predictive or prognostic implications (35). It has recently been suggested that the expression changes of genes affected by K-RAS mutations have higher prognostic relevance than the primary genetic alteration. So, secondary effects of a mutation are prominent and can be used as a prognostic signature in lung cancer (36). Therefore, the expression changes of the K-RAS gene and protein in the peripheral and blood samples were investigated in this study. Since it has been recently shown that oncogenic K-RAS induces lung tumorigenesis through different molecular mechanisms, such as miRNA modulation (7), the expression and existence of 3 aforementioned miRNAs miRNA-128, miR-335-5p, and miR-1254 were also investigated. MiRNA-30c and miRNA-21 levels have been shown to significantly elevate in NSCLC patients’ tissues at early stages compared to normal lung tissues and in plasma from the same patients.
Another major finding of the present study was the higher mRNA expression level of VEGF and the lower expression of miR-128b in the peripheral blood and tissue samples of NSCLC patients compared to healthy subjects. Unlike miRNA-128b, both VEGF protein and mRNA were also detected in more people with NSCLC than in healthy subjects. These findings suggest that VEGF can be considered a serum and tissue biomarker in patients with NSCLC (9). This study reported that the expression level of miR-128 is significantly downregulated in NSCLC tissues and cells and is significantly correlated with NSCLC pathological grading and lymph node metastasis. These findings showed that down-regulation of miR-128 may inhibit VEGF expression in NSCLC tissue and peripheral blood. The over-expression of miR-128 has been shown to inhibit VEGF-C expression in NSCLC cells (9).
5.1. Conclusions
The data obtained from the current study show that miR-335-5p, miR-1254, and miR-128 can play a role in NSCLC pathogenesis and tumorigenesis, at least in part, by modulation of proliferation, migration, and angiogenesis via targeting VEGF and K-RAS signaling pathways. K-RAS and VEGF may be target genes for miR-335-5p, miR-1254, and miR-128 in NSCLC patients.