1. Background
Liver cancer, particularly hepatocellular carcinoma (HCC), stands as one of the most prevalent and lethal malignancies worldwide (1). Despite recent advancements in diagnosis and treatment, the precise molecular mechanisms involved in HCC progression remain incompletely understood (2). Recently, the role of non-coding RNAs (ncRNAs) in regulating gene expression and cancer progression has garnered significant attention (3). Among various signaling pathways, the Hippo pathway plays a pivotal role in regulating cell growth and proliferation in HCC (4).
The Hippo signaling pathway, through regulation of YAP and TAZ transcription factor activity, plays a vital role in controlling organ size and tissue homeostasis (4). Dysregulation of this pathway is closely associated with HCC progression (5). Furthermore, recent studies have shown that ncRNAs, including lncRNAs and circRNAs, can play important roles in HCC pathogenesis by regulating the expression of key genes in the Hippo pathway and other HCC pathways (6).
MicroRNAs (miRNAs) have emerged as crucial regulators in various cancer types, including HCC. These small ncRNAs play significant roles in post-transcriptional regulation of gene expression and have been implicated in multiple aspects of hepatocarcinogenesis, including cell proliferation, apoptosis, and metastasis (3). The HCC pathway involves intricate interactions among diverse signaling cascades, transcription factors, and regulatory RNAs. Key genes in this pathway, such as PRKCB and SMARCA2, are involved in HCC progression and may be potential targets for therapeutic interventions (7). Understanding the interplay between miRNAs, other ncRNAs, and mRNAs in the context of the HCC pathway is crucial for unraveling the molecular mechanisms underlying liver cancer development and progression.
2. Objectives
The main objectives of this study are:
- To investigate the interactions between key genes involved in the Hippo pathway (LEF1, MOB1A) and HCC pathways (PRKCB and SMARCA2), as well as specific ncRNAs and miRNAs.
- To analyze gene expression patterns in HCC through bioinformatic analysis of two independent tissue datasets.
- To identify candidate ncRNAs and miRNAs with potential interactions with these genes using computational predictions and a comprehensive literature review (6).
- To examine the complex network of interactions between various RNA types (mRNAs, lncRNAs, circRNAs, and miRNAs) in the HCC (8).
- To focus on two specific ncRNAs: RN7SL1 (lncRNA) and hsa_circ_0001380 (circRNA) (9), investigating their potential roles in regulating gene expression and signaling pathways related to HCC (10).
- To validate computational findings by experimentally testing the expression of selected ncRNAs in HCC (HepG2) and normal skin (fibroblast) cell lines.
Through this multi-faceted approach combining in silico analyses and experimental validation, we aim to provide a more comprehensive understanding of the regulatory mechanisms in HCC and potentially identify novel therapeutic targets (11).
3. Methods
3.1. Gene and Non-coding RNAs Selection
Signaling Pathway Genes: The genes involved in the Hippo signaling pathway (LEF1, MOB1A) and the HCC pathway (PRKCB, SMARCA2) were selected based on their established roles in liver cancer progression (12, 13).
ncRNAs: ncRNAs (RN7SL1, hsa_circ_0001380) were chosen for this study based on their potential involvement in various cancer types and HCC as reported in previous studies (12, 14).
3.2. Differential Expression Analysis
Differential expression analysis was performed, using two datasets (GSE14520, platforms GPL3921, and GPL571) obtained from the Gene Expression Omnibus (GEO) database (15). The analysis was conducted, using the GEO2R tool, which employs the limma R package (16). This analysis was specifically performed to examine the expression patterns of pre-selected genes in HCC samples compared to normal liver tissues. The expression changes were quantified, using log2 fold change values, with an adjusted P-value < 0.05 considered statistically significant. This approach allowed us to characterize the expression profiles of genes of interest in the context of HCC.
3.3. Sequence Retrieval
3.3.1. Long Non-coding RNA (lncRNA) Sequences
The sequences of the selected lncRNAs RN7SL1 were obtained from the LncPedia database (17). For RN7SL1, all available transcript variants were downloaded in FASTA format to ensure a comprehensive analysis of potential functional transcripts.
3.3.2. Circular RNA Sequence
The sequence of the selected circRNA (hsa_circ_0001380) was retrieved from the CircBank database (18). The sequence was downloaded in FASTA format for subsequent analysis.
3.3.3. mRNA Sequences
For the selected genes involved in the Hippo signaling pathway (LEF1, MOB1A) and the HCC pathway (PRKCB, SMARCA2), all known transcript variants were retrieved from the National Center for Biotechnology Information (NCBI) database (10). The sequences were downloaded in FASTA format, allowing for a thorough examination of potential interactions between these mRNAs and the selected ncRNAs.
3.3.4. miRNA Prediction and Analysis
To further investigate the potential regulatory mechanisms involving the selected genes, a comprehensive miRNA analysis was performed:
- miRNA Target Prediction: The 3'UTR and 5'UTR regions of the selected genes (LEF1, MOB1A, PRKCB, SMARCA2) were analyzed, using the miRWalk database (19). The search was filtered to include only miRNAs validated in the miRTarBase.
- miRNA Sequence Retrieval: The sequences of the identified miRNAs were obtained from the miRBase database (20).
- Conversion to Gene Sequences: The miRNA sequences were converted to their corresponding gene sequences for further analysis.
- Integration with Physical Interaction Analysis: The gene sequences of the identified miRNAs were included in the physical interaction prediction analysis alongside the previously selected ncRNAs and mRNAs.
This additional step facilitated a more thorough examination of potential regulatory interactions, encompassing both direct ncRNA-mRNA interactions and potential miRNA-mediated regulation.
3.4. Physical Interaction Prediction
The potential physical interactions between the selected mRNAs and ncRNAs were predicted, using the long non-coding RNA-target analysis resource (LncTAR) tool (21). Long non-coding RNA-target analysis resource is designed to identify putative interactions between long ncRNAs and their target RNAs based on complementary base pairing and thermodynamic stability.
The FASTA sequences of all transcript variants for both mRNAs and ncRNAs, obtained as described in the previous section, were used as input for LncTAR. The analysis was performed with the following parameters:
- Minimum free energy (MFE) threshold: -15 kcal/mol
These parameters were chosen based on recommendations in the literature and previous studies using LncTAR for similar analyses (22). The tool generated a list of potential interactions between the input sequences, ranked by their predicted binding strength and stability.
3.5. Cell Culture and Gene Expression Analysis
3.5.1. Cell Culture
To validate the computational analyses, cell lines derived from liver cancer (HepG2) and normal fibroblast (NIH) were used for laboratory experiments. The HepG2 liver cancer cell line with code BN_0012.1.10 and the normal fibroblast cell line NIH with code BN_0012.1.21 were obtained as cultured cells from the Bon Yakhteh Research Center (Bon Yakhteh, Iran).
3.6. RNA Extraction, cDNA Synthesis, and RT-qPCR
Total RNA was extracted from cultured cells, using TRIzol (Sigma-Aldrich, Germany). The quality of RNA was assessed, using RNase-free DNase I (Sinaclon, IRAN). cDNA synthesis was performed, using a cDNA synthesis kit (Biofact, Korea). RT-qPCR was carried out, using Roche real-time PCR systems. Each reaction consisted of 10 pmol/µL of each primer, 10 µL SYBR Green PCR Master Mix (Takara, Japan), and 50 ng cDNA in a final volume of 20 µL, following the protocol. The expression levels were normalized, using β-actin.
The specific primer sequences for the candidate genes were designed, using Beacon Designer Version 7.2 (USA) and ordered through Metabion Company (Germany). The primer specificity was confirmed, using "NCBI Primer-BLAST". The primer sequences are listed in Table 1.
| Name | Sequences (5'-3') | TM (°C) | Size of Gene | Product Size (bp) | Per 13 µL (Total Volume) |
|---|---|---|---|---|---|
| lnc-LRR1-1 | 60 | 300 | 102 | 1 | |
| F | GGCTGAGGCTGGAGGAT | ||||
| R | GGGAGGTCACCATATTGATG | ||||
| hsa_circ_0001380 | 60 | 247 | 87 | 1 | |
| F | GGAACCAGAACCATTATTTG | ||||
| R | TCTGCAAGGGTAGTTAATTTC | ||||
| Beta-actin | 60 | 5589 | 144 | 1 | |
| F | CCTGGCACCCAGCACAAT | ||||
| R | GGGCCGGACTCGTCATAC |
a All measurements were carried out in duplicate. We report the data using the ΔΔCt method.
b lnc-LRR1-1 primers detect transcripts lnc-LRR1-1:1 and lnc-LRR1-1:2.
Primers for RN7SL1: The primers were designed to detect both transcripts lnc-LRR1-1:1 and lnc-LRR1-1:2, which exhibited 99% similarity.
Sequence similarities were analyzed, using the BLAST tool (23) available on the NCBI website.
In response to concerns regarding the specificity of the primers used for lnc-LRR1-1 and hsa_circ_0001380, we conducted a thorough analysis to confirm specificity. The lnc-LRR1-1 transcript is an isoform of the RN7SL1 gene. Using the UCSC Genome Browser PCR tool, we observed that our designed primers amplified 9 RN7SL1 isoforms with over 90% sequence similarity, confirming that the primers target different isoforms of the same gene rather than nonspecific off-target sequences.
For hsa_circ_0001380, we verified specificity with UCSC In-Silico PCR, which showed alignment with the host gene UBXN7, from which the circRNA originates, not unrelated genes. Thus, the primers amplify the circRNA's genomic sequence as expected.
Additionally, melting curve analysis from our real-time PCR experiments produced single sharp peaks for both genes (Tm: ~86.32°C and ~81.7°C), indicating specific amplification without nonspecific products or primer-dimers. The single-peaked melting curves support our primer specificity and the reliability of the gene expression data. The melting curve plots (Appendix 1 in Supplementary File) are included as evidence of primer specificity. Appendix 1 in Supplementary File illustrates the single-peaked melting curves for hsa_circ_0001380 and lnc-LRR1-1, confirming the specificity of the primers used.
Gene expression levels were calculated, using the 2-ΔΔCt method, with β-actin as the reference gene. All experiments were performed in duplicate.
3.7. Statistical Analysis
Statistical analyses for the gene expression data were performed, using the Relative Expression Software Tool (REST) 2009 (24). Relative Expression Software Tool employs a mathematical model that calculates the relative expression ratio based on the PCR efficiencies and the mean crossing point deviation between the sample and control groups. This tool incorporates a statistical test to determine the significance of the expression ratios, which accounts for issues with amplification efficiency and reference gene normalization.
The expression levels of the target genes were normalized to β-actin as the reference gene. The software performs randomization tests (10,000 randomizations) to determine the statistical significance of the calculated expression ratios. A P-value < 0.05 was considered statistically significant for all analyses.
For the bioinformatics analyses, including the differential expression analysis of the GEO datasets, the Benjamini-Hochberg procedure was used to control the false discovery rate (FDR) in multiple tests. An adjusted P-value < 0.05 was considered statistically significant.
4. Results
4.1. Differential Gene Expression Analysis in Hepatocellular Carcinoma
Gene expression patterns in HCC were investigated, using two independent platforms (GPL3921 and GPL571) from the GSE14520 dataset, obtained from the Gene Expression Omnibus (GEO) database. The GPL3921 platform encompassed 225 tumor samples and 220 non-tumor samples, facilitating a comprehensive analysis of gene expression variations. The GPL571 platform, though smaller in scale, provided additional validation with 22 tumor samples and 21 normal samples. This dual-platform approach facilitated a comprehensive and robust analysis of gene expression in HCC. The analysis investigated the overall gene expression alterations and specifically concentrated on genes involved in the Hippo signaling pathway (LEF1, MOB1A) and genes associated with the HCC pathway (PRKCB, SMARCA2). The results of these analyses are presented in Tables 2 and 3.
| Gene Symbol | LogFC | Adjusted P-Value |
|---|---|---|
| LEF1 | 0.7068694 | 5.44e-19 |
| MOB1A | 0.1175415 | 8.29e-04 |
| PRKCB | -0.1233407 | 2.02e-03 |
| SMARCA2 | -0.0180284 | 2.99e-01 |
| Gene Symbol | LogFC | Adjusted P-Value |
|---|---|---|
| LEF1 | 0.61145671 | 9.57e-04 |
| MOB1A | 0.03974242 | 5.95e-01 |
| PRKCB | -0.19636147 | 6.70e-02 |
| SMARCA2 | -0.09621861 | 1.52e-01 |
4.2. Global Gene Expression Changes
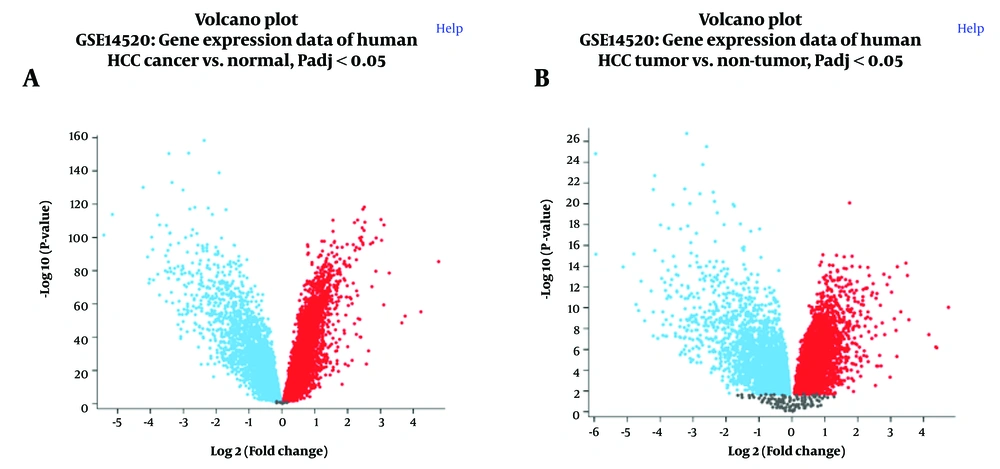
Volcano plots generated from both platforms revealed substantial changes in gene expression between tumor and non-tumor samples (Figure 1A and B). A symmetrical distribution of up-and down-regulated genes was observed, with a substantial number of genes exhibiting significant differential expression (adjusted P-value < 0.05). The volcano plots from both platforms demonstrated similar patterns, indicating consistency in the overall gene expression changes detected.
Volcano plots depicting differential gene expression in hepatocellular carcinoma (HCC) versus normal liver tissue. A, GPL3921 platform; and B, GPL571 platform from the GSE14520 dataset. The X-axis represents log2 fold change, and the y-axis represents -log10 (adjusted P-value). Red dots indicate significantly upregulated genes, blue dots indicate significantly downregulated genes (adjusted P-value < 0.05), and gray dots represent genes with non-significant changes.
4.3. Sample Clustering and Expression Distribution
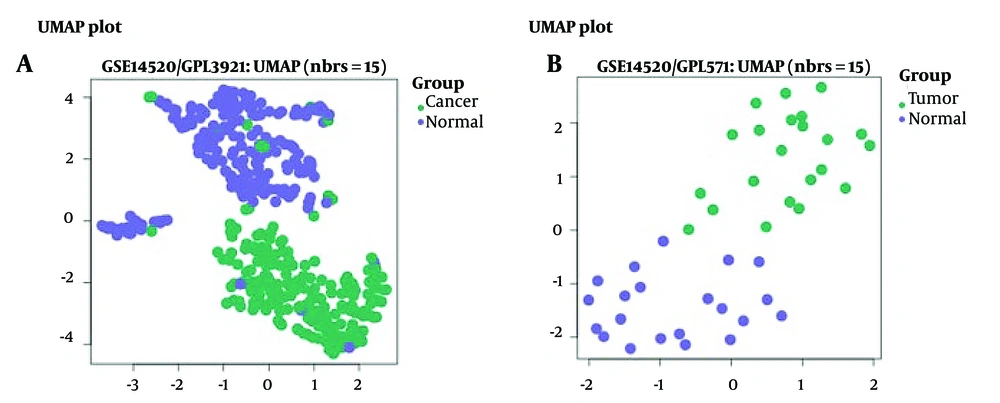
Uniform manifold approximation and projection (UMAP) analysis was performed to visualize the clustering of tumor and normal samples based on their gene expression profiles (Figure 2A for GPL3921 and Figure 2B for GPL571). Both platforms demonstrated clear separation between tumor (green) and normal (purple) samples, indicating distinct gene expression profiles between these two groups. This separation supports the reliability of the dataset and the significance of the observed differential gene expression.
Uniform Manifold Approximation and Projection (UMAP) plots show distinct clustering of hepatocellular carcinoma (HCC) and normal liver tissue samples. A, GPL3921 platform; and B, GPL571 platform from the GSE14520 dataset. Green dots represent tumor samples, and purple dots represent normal tissue samples. The clear separation between clusters demonstrates distinct gene expression profiles in HCC versus normal liver tissue.
4.4. Analysis of Selected Genes
The expression changes of the selected genes in both platforms are summarized in Tables 2 and 3.
Analysis of the selected genes revealed varying expression patterns across the two platforms:
4.5. Hippo Signaling Pathway-Related Genes
LEF1 showed significant upregulation in both platforms (adj. P-value < 0.001 for GPL3921 and adj. P-value < 0.01 for GPL571).
MOB1A displayed significant upregulation in GPL3921 (adj. P-value < 0.01) but not in GPL571 (adj. P-value = 0.595).
4.6. Hepatocellular Carcinoma Pathway-Related Genes
PRKCB demonstrated downregulation in both platforms, although it was only statistically significant in GPL3921 (adj. P-value <0.01).
SMARCA2 exhibited a significant downregulation in both platforms, although statistical significance was not attained in either (adjusted P-value for GPL3921: 0.299, adjusted P-value for GPL571: 0.152).
The variations observed between the two platforms highlight the importance of using multiple datasets in expression analysis. Despite these differences, the overall trends of gene expression changes provide a foundation for further investigation of their potential roles in HCC progression.
4.7. Physical Interaction Prediction
Physical interactions between selected ncRNAs and mRNAs in the context of HCC were predicted, using the LncTAR tool. A total of 19 potential interactions were identified (Table 4), involving multiple transcript isoforms of ncRNAs (RN7SL1 [represented by lnc-LRR1-1:1 and lnc-LRR1-1:2] and hsa_circ_0001380) and 4 mRNAs (LEF1, MOB1A, PRKCB, and SMARCA2). Interactions were filtered based on a normalized ΔG (ndG) threshold of -0.15.
| ncRNA | Target Gene | Transcript ID | dG (kcal/mol) | ndG | ncRNA Position | Target Position |
|---|---|---|---|---|---|---|
| lnc-LRR1-1:1 | LEF1 | NM_001130713.3 | -47.29 | -0.1636 | 1-300 | 720-1019 |
| lnc-LRR1-1:2 | LEF1 | NM_001130713.3 | -47.29 | -0.1636 | 1-297 | 721-1017 |
| lnc-LRR1-1:1 | LEF1 | NM_001130714.3 | -47.29 | -0.1636 | 1-300 | 720-1019 |
| lnc-LRR1-1:2 | LEF1 | NM_001130714.3 | -47.29 | -0.1636 | 1-297 | 721-1017 |
| lnc-LRR1-1:1 | LEF1 | NM_016269.5 | -47.29 | -0.1636 | 1-300 | 720-1019 |
| lnc-LRR1-1:2 | LEF1 | NM_016269.5 | -47.29 | -0.1636 | 1-297 | 721-1017 |
| lnc-LRR1-1:1 | MOB1A | NM_001317111.2 | -57.29 | -0.1916 | 1-300 | 3226-3525 |
| lnc-LRR1-1:2 | MOB1A | NM_001317111.2 | -59.01 | -0.2130 | 1-297 | 3228-3524 |
| lnc-LRR1-1:1 | MOB1A | NM_001317110.2 | -57.29 | -0.1916 | 1-300 | 3242-3541 |
| lnc-LRR1-1:2 | MOB1A | NM_001317110.2 | -59.01 | -0.2130 | 1-297 | 3244-3540 |
| lnc-LRR1-1:1 | MOB1A | NM_001317112.2 | -57.29 | -0.1916 | 1-300 | 3081-3380 |
| lnc-LRR1-1:2 | MOB1A | NM_001317112.2 | -59.01 | -0.2130 | 1-297 | 3083-3379 |
| lnc-LRR1-1:1 | MOB1A | NM_018221.5 | -57.29 | -0.1916 | 1-300 | 3245-3544 |
| lnc-LRR1-1:2 | MOB1A | NM_018221.5 | -59.01 | -0.2130 | 1-297 | 3247-3543 |
| lnc-LRR1-1:1 | PRKCB | NM_212535.3 | -45.54 | -0.1621 | 1-300 | 57-356 |
| lnc-LRR1-1:2 | PRKCB | NM_212535.3 | -43.61 | -0.1552 | 1-297 | 59-355 |
| lnc-LRR1-1:1 | PRKCB | NM_002738.7 | -45.54 | -0.1621 | 1-300 | 57-356 |
| lnc-LRR1-1:2 | PRKCB | NM_002738.7 | -43.61 | -0.1552 | 1-297 | 59-355 |
| hsa_circ_0001380 | SMARCA2 | NM_001289399.2 | -13.25 | -0.1721 | 126-247 | 1-122 |
The predicted interaction sites for LEF1 and PRKCB were observed to be located in the middle region of the target mRNA transcripts, while for MOB1A, the interactions were predominantly near the 3' end of the transcripts.
The strongest predicted interactions were identified between lnc-LRR1-1:2 and several MOB1A transcript variants, with dG = -59.01 kcal/mol and ndG = -0.2130. These interactions were observed to encompass the entire length of the lnc-LRR1-1:2 sequence (positions 1-297).
A specific interaction between the circular RNA hsa_circ_0001380 and SMARCA2 was detected, although with a lower interaction strength compared to the interactions between lncRNAs. This interaction was localized at the 5' end of the SMARCA2 transcript, spanning positions 1-122 on the mRNA and involving positions 126-247 of hsa_circ_0001380.
Multiple isoforms of LEF1 and MOB1A were found to interact with both lnc-LRR1-1:1 and lnc-LRR1-1:2, with consistent interaction strengths for each gene. For LEF1, all interactions showed dG = -47.29 kcal/mol and ndG = -0.1636, while for MOB1A, lnc-LRR1-1:1 interactions had dG = -57.29 kcal/mol and ndG = -0.1916, and lnc-LRR1-1:2 interactions had dG = -59.01 kcal/mol and ndG = -0.2130.
For PRKCB, interactions were predicted with both lnc-LRR1-1:1 and lnc-LRR1-1:2 on two transcript variants. The interactions with lnc-LRR1-1:1 showed identical strengths (dG = -45.54 kcal/mol, ndG = -0.1621) for both transcripts, while interactions with lnc-LRR1-1:2 were slightly weaker (dG = -43.61 kcal/mol, ndG = -0.1552).
These computational predictions provide a basis for further experimental investigations into the potential regulatory roles of these ncRNAs in HCC-related pathways. The consistency in interaction strengths across multiple transcript variants for each gene suggests potentially important functional relationships between these ncRNAs and their target mRNAs.
4.8. miRNA Interaction Analysis
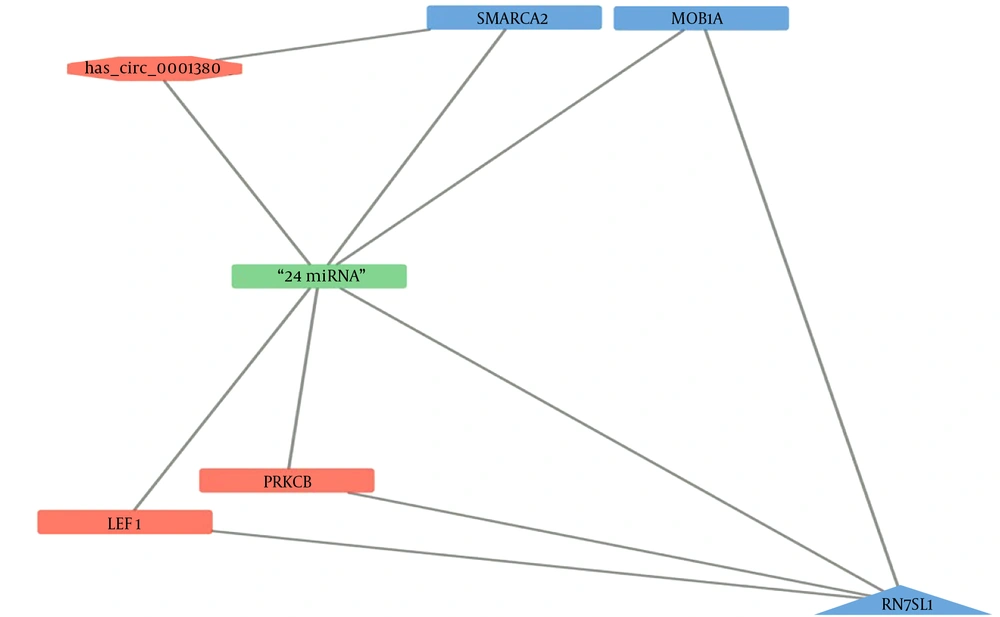
Physical interactions between the selected ncRNAs, mRNAs, and miRNAs in the context of HCC were predicted, using the LncTAR tool. The analysis revealed a complex network of potential interactions involving 24 miRNAs, the ncRNAs (RN7SL1 represented by lnc-LRR1-1:1 and lnc-LRR1-1:2, and hsa_circ_0001380), and the mRNAs of LEF1, MOB1A, PRKCB, and SMARCA2. The key findings are summarized below:
4.8.1. miRNA Interactions with ncRNAs
- hsa_circ_0001380 showed potential interactions with all 24 miRNAs, with interaction strengths (dG) ranging from -3.07 to -9.24 kcal/mol.
- Both lnc-LRR1-1:1 and lnc-LRR1-1:2 demonstrated interactions with all 24 miRNAs, with dG values ranging from -3.17 to -8.78 kcal/mol.
4.8.2. miRNA Interactions with mRNAs
- LEF1: Interactions were predicted across multiple transcript variants (NM_001130713.3, NM_001130714.3, NM_001166119.2, NM_016269.5) with dG values ranging from -4.86 to -11.98 kcal/mol.
- MOB1A: Various transcript variants (NM_001317111.2, NM_001317110.2, NM_001317112.2, NM_018221.5) showed interactions with dG values between -3.17 and -11.98 kcal/mol.
- PRKCB: Two transcript variants (NM_212535.3, NM_002738.7) demonstrated interactions with dG values from -4.00 to -10.68 kcal/mol.
- SMARCA2: Multiple transcript variants (NM_001289399.2, NM_001289400.2, NM_001289396.2, NM_139045.4, NM_001289397.2, NM_001289398.2, NM_003070.5) showed interactions with dG values ranging from -4.86 to -12.29 kcal/mol.
4.8.3. Interaction Patterns
- The majority of predicted interactions for both ncRNAs and mRNAs extended nearly the entire length of the miRNA sequences (typically positions 1-22).
- Interaction sites on mRNAs were distributed across various regions, including 5' UTR, coding sequences, and 3' UTR.
4.8.4. Strongest Interactions
- The strongest interaction for hsa_circ_0001380 was with hsa-miR-193b-3p (dG = -9.24 kcal/mol).
- For lnc-LRR1-1:1 and lnc-LRR1-1:2, the strongest interaction was with hsa-miR-564 (dG = -8.78 kcal/mol).
- Among mRNAs, SMARCA2 showed the strongest predicted interaction with hsa-miR-4742-3p (dG = -12.29 kcal/mol).
These results suggest a complex regulatory network involving miRNAs, ncRNAs, and mRNAs in HCC, with potential implications for gene expression regulation and disease progression.
Figure 3 presents a network visualization of these interactions, created using Cytoscape (25). In this representation, the 24 miRNAs are depicted as a single node for simplification, given the consistent pattern of interactions across all analyzed RNAs.
4.9. Experimental Validation of ncRNA Expression
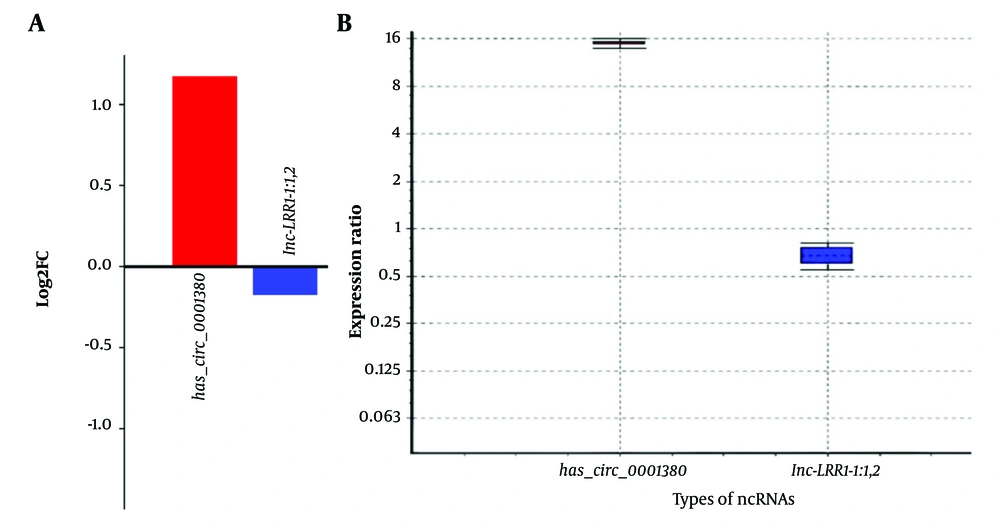
Quantitative PCR (qPCR) analysis was performed to validate the expression of selected ncRNAs in HepG2 liver cancer cells compared to normal skin fibroblasts. The analysis included circRNA (hsa_circ_0001380) and lncRNAs (lnc-LRR1-1:1,2), with β-actin serving as a reference gene. The results, analyzed using the REST, revealed significant expression changes for all tested ncRNAs (P < 0.001) (Figure 4A, B and Table 5).
| Gene | Type | Reaction Efficiency | Expression | Std. Error | 95% C.I. | P (H1) | Result |
|---|---|---|---|---|---|---|---|
| β-actin | REF | 1.0 | 1.000 | ||||
| hsa_circ_0001380 | TRG | 1.0 | 14.933 | 14.236 - 15.673 | 13.951 - 15.986 | 0.000 | Upregulation |
| lnc-LRR1-1:1,2 | TRG | 1.0 | 0.670 | 0.574 - 0.783 | 0.556 - 0.809 | 0.000 | Downregulation |
These results suggest differential expression of these ncRNAs between HCC and normal cell lines, indicating their potential involvement in HCC-related cellular processes.
Expression analysis of non-coding RNAs hsa_circ_0001380 and lnc-LRR1-1:1,2 in HCC cell line compared to normal fibroblasts; A, Log2 fold change (log2FC) of hsa_circ_0001380 and lnc-LRR1-1:1,2 expression in HepG2 (HCC cell line) compared to NIH (normal fibroblast cell line). The bar graph shows the upregulation of hsa_circ_0001380 (log2FC ≈ 1.2) and downregulation of lnc-LRR1-1:1,2 (log2FC ≈ -0.2) in HepG2 cells; B, Box plot representing the expression ratio of hsa_circ_0001380 and lnc-LRR1-1:1,2 in HepG2 cells relative to NIH cells. The y-axis shows the expression ratio on a log2 scale. hsa_circ_0001380 exhibits significantly higher expression (median ≈ 16-fold increase), while lnc-LRR1-1:1,2 shows slightly lower expression (median ≈ 0.7-fold change) in HepG2 compared to NIH cells.
5. Discussion
This study investigated the intricate regulatory networks involving ncRNAs and key genes in the Hippo signaling and HCC pathways, revealing significant insights into the molecular mechanisms underlying HCC progression.
Expression analysis revealed significant upregulation of LEF1 in HCC samples, consistent with its potential oncogenic role in liver cancer (26). Conversely, PRKCB showed consistent downregulation, suggesting a possible tumor-suppressive function. The variable expression patterns of MOB1A and SMARCA2 highlight the complexity of signaling regulation in HCC.
Computational predictions identified numerous potential interactions between the selected ncRNAs (lnc-LRR1-1:1, lnc-LRR1-1:2, and hsa_circ_0001380) and mRNAs of key HCC-related genes. Notably, strong interactions were predicted between lnc-LRR1-1:2 and various MOB1A transcript variants, suggesting a potential regulatory role for this lncRNA in Hippo pathway signaling. These interactions were predominantly observed in the 3' UTR regions of the target mRNAs, indicating possible post-transcriptional regulation.
The interaction between lnc-LRR1-1:1/2 and LEF1 transcripts may indicate a regulatory mechanism influencing Wnt/β-catenin signaling in the HCC (26). Interestingly, hsa_circ_0001380 showed a distinct interaction pattern with SMARCA2, localized at the 5' end of the transcript, suggesting a potential role in transcriptional regulation.
Experimental validation confirmed differential expression of hsa_circ_0001380 and lnc-LRR1-1:1,2 in HCC cell lines. The significant upregulation of hsa_circ_0001380 aligns with its predicted interactions and suggests a potential oncogenic role. The slight downregulation of lnc-LRR1-1:1,2 indicates a more complex regulatory landscape.
The complex network of interactions involving miRNAs, ncRNAs, and mRNAs suggests potential competing endogenous RNA (ceRNA) mechanisms in HCC (27). The most potent predicted interaction between hsa_circ_0001380 and hsa-miR-193b-3p may represent a novel regulatory pathway in the progression of HCC.
While this study provides valuable insights into the role of ncRNAs in HCC, it is important to consider its limitations. The in-silico predictions, although robust, require further experimental validation in diverse HCC cell lines and patient-derived samples (28). Our findings on lnc-LRR1-1:1/2 and hsa_circ_0001380 align with recent studies that have highlighted the importance of ncRNAs in HCC progression. For instance, Zhang et al. demonstrated that circRNA-104075 promotes HCC progression through a ceRNA mechanism involving miR-582-3p (29). Our study extends these findings by providing a comprehensive analysis of ncRNA interactions with key genes in the Hippo and HCC pathways, offering a novel perspective on the regulatory landscape of HCC. Future studies should focus on elucidating the specific molecular mechanisms, by which these ncRNAs modulate gene expression in HCC, potentially through CRISPR-Cas9 mediated knockout or overexpression experiments (3).