1. Background
Breast cancer, a disease of profound significance, stands as a global health challenge affecting millions of people in the world (1). Its prevalence and impact on individuals and communities underscore the pivotal role of understanding, addressing, and combating this formidable adversary. Using molecular and histological data, breast cancer can be classified into 3 main subtypes: Hormone receptor-positive breast cancer [expressing estrogen receptor (ER+) or progesterone receptor (PR+)], human epidermal growth factor receptor 2-positive breast cancer (HER2+), and triple-negative breast cancer (lacking expression of ER, PR, and HER2). Changes in different cellular pathways and biological processes (BPs) of the cell can be the main cause of breast cancer (2, 3).
The Wnt pathway, a critical cell growth and development regulator, is significantly implicated in breast cancer. Disruption of this pathway contributes significantly to the pivotal role in the initiation and progression of breast cancer, fostering uncontrolled cell proliferation. Interactions between the Wnt pathway and other signaling pathways amplify its impact on breast cancer, influencing tumor development and metastasis (4). Targeting the Wnt pathway emerges as a promising therapeutic strategy to impede breast cancer cell growth and spread. Additionally, Wnt signaling’s involvement in maintaining cancer stem cells, driving tumor initiation, recurrence, and therapy resistance, adds complexity to breast cancer biology (4). Crosstalk with hormone receptors, like estrogen receptors, further complicates the role of the Wnt pathway in breast cancer. This intricate relationship requires thorough exploration for the identification of biomarkers and the development of targeted therapies. Ongoing research efforts aim at unraveling the precise mechanisms underlying Wnt pathway influence in breast cancer, paving the way for more effective and personalized treatment approaches (5).
Network analysis is a pivotal tool in cancer treatment, offering a multifaceted approach to understanding and combating this complex disease. By delving into intricate interactions between genes, proteins, and pathways, network analysis aids in the identification of crucial biomarkers and therapeutic targets tailored to specific cancer types. This knowledge proves invaluable in the era of personalized medicine, allowing for more precise and effective treatment strategies. Moreover, network analysis is a key factor in deciphering the heterogeneous nature of tumors. This comprehensive understanding is essential for developing targeted therapies that consider the diverse characteristics of individual cancers (6). Crucially, network analysis unravels resistance mechanisms, shedding light on how cancers adapt and evade treatments (7). Armed with this knowledge, researchers can develop strategies to overcome resistance, a significant challenge in cancer therapy. Finally, network analysis fosters collaboration among researchers, clinicians, and bioinformaticians. By sharing data and insights, the scientific community can collectively advance our understanding of cancer biology and improve treatment strategies, ultimately bringing us closer to more effective and targeted cancer therapies (7).
MicroRNAs (miRNAs) are small, naturally occurring non-coding RNA molecules that play a vital role in controlling gene expression. Robust evidence indicates the disruption of pertaining to miRNA levels in cancers affecting humans, stemming from diverse processes including miRNA gene duplication as well as removal, combined with irregular transcriptional activity, disturbed epigenetic modifications, and flaws during miRNA synthesis. These dysregulated miRNAs can act as either tumor-promoting genes or tumor-restraining genes, influencing essential neoplasm characteristics like continuous proliferation pathways, evasion of expansion suppressors, avoidance of programmed cell death, initiation of tissue invasion and cancer dissemination, and stimulation of the formation of new blood vessels. An expanding body of research underscores miRNAs as promising indicators for cancer detection, prognosis, and therapeutic targets, necessitating further exploration and validation (8, 9).
The present study focuses on understanding the disruption in miRNA expression, pinpointing central genes, and uncovering the complex interactions between signaling pathways. By applying clustering techniques, we analyze the functional components within these networks, offering a comprehensive perspective on the interactions between critical molecular components. Additionally, a thorough investigation of promoter regions associated with hub genes highlights the regulatory components of transcription that propel breast cancer development and progression.
2. Objectives
By integrating these multiple approaches, our research aims to provide detailed insights into the molecular framework of breast carcinoma, potentially informing diagnostic approaches and the identification of targets for therapy. While investigating the intricacies of miRNAs control networks, this examination signifies a significant step toward deciphering the molecular complexity that characterizes breast carcinoma.
3. Methods
3.1. Exploration of Protein Interactions Network
To compile the list of miRNAs that regulate the Wnt cellular signaling cascade, we referenced an article reviewed by Rahmani et al. (10). The complex network of protein-protein interactions (PPIs) within this pathway was, then, unraveled using the predictive power of the STRING database (version 11.5). This resource incorporates both physical (direct) and functional (indirect) interactions, amalgamating insights from computational tools, inter-species knowledge sharing, and data extracted from key databases (11).
For a visual representation and identification of central players within the PPI network, Cytoscape software (version 3.9.1) was employed. The CytoHubba (version 0.1) facilitated the recognition of hub proteins, essential for coordinating network activity, employing 11 topological analysis methods (12).
3.2. Comprehensive Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Pathway Enrichment Evaluation
To meticulously assess gene functions, the STRING tool variant 11.5 was utilized for functional annotation using gene ontology (GO). This unraveled molecular functions (MFs), BPs, and cellular components (CC). Simultaneously, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was executed, aiming to explore the linked molecular networks and cellular signaling cascade associated with the studied genes (12).
3.3. Examination of Network Clusters
To decipher the structural principles of the network, CytoCluster (version 2.1.0) was employed, utilizing the IPCA approach for identifying protein complexes with a rigorous threshold of 2. STRING software (variant 11.5) was subsequently used to analyze the genomic profile of each cluster, providing insights into the intricate associations between genes and specific KEGG pathways (13).
3.4. Analysis of Regulatory Motifs in Hub Gene Promoters
To clarify the regulatory mechanisms, controlling hub genes and their upstream flanking regions (UFRs) were meticulously extracted from the Ensembl database. MEME Suite (version 5.5.2) was, then, employed for the identification of conserved motifs within these genomic sequences. The Tomtom software (version 5.5.2) cross-referenced against the HOCOMOCO Human (v11 CORE), unveiling known cis-regulatory elements. Additionally, using the GoMo tool, we identified potential functional roles of the discovered motifs. Through this detailed analysis, we aim to decipher the sophisticated regulatory language within the UFRs of hub genes, shedding light on the complex molecular processes that regulate gene expression (14, 15).
4. Results
4.1. Identifying MicroRNAs Regulating the Wnt/β-Catenin Cascade in Breast Cancer Patients
The miRNAs and their target genes were compiled from an article reviewed by Rahmani et al. (10) and information available in the miRDB database. Table 1 provides an extensive catalog of miRNAs regulating the Wnt/β-catenin cascade in individuals with breast cancer. After identifying aberrantly regulated miRNAs targeting the Wnt cascade from prior group research, we pinpointed their target genes, focusing on those with a miRDB database score above 95.
| Targets | Molecular Alterations | Functions |
|---|---|---|
| GSK-3 β, APC, and ICAT | ||
| miR-1229 | Upregulation | Inducing cancer growth and growth |
| ICAT | ||
| miR-1301 | Upregulation | Induces tumor cell proliferation |
| APC | ||
| miR-125b | Upregulation | Inducing cell growth and metastasis |
| GSK-3 β | Upregulation | Inducing drug resistance |
| miR-3646 | ||
| miR-29a | ||
| PTEN | ||
| miR-301 | Upregulation | Inducing cell growth and metastasis |
| WIF1 and Wnt5a | ||
| miR-374a | Upregulation | Inducing tumor cell metastasis |
| DACT3 | ||
| miR-638 | Upregulation | Inducing cell proliferation |
| β-catenin | ||
| miR-373 | Upregulation | Promoting EMT |
| β-catenin | Downregulation | Inhibiting tumor cell proliferation |
| miR-26b | ||
| miR-135 | ||
| miR-214 | ||
| miR-216a | ||
| miR-340 | ||
| UBE3C | ||
| miR-30a-5p | Downregulation | Inhibiting tumor cell proliferation and metastasis |
| CREPT | Downregulation | Inhibiting tumor growth and metastasis |
| miR-138 | ||
| miR-449b-5p | ||
| ACVR1 | ||
| miR-384 | Downregulation | Inhibiting tumor metastasis |
| CTHRC1 | ||
| miR-30 | Downregulation | Inhibiting cancer cell invasion and metastasis |
| Wnt1 | ||
| miR-140-5p | Downregulation | Inhibiting tumor cell proliferation |
| Frizzled 5 | ||
| miR-224 | Downregulation | Inhibiting tumor progression |
| Frizzled 7 | ||
| miR-1 | Downregulation | Inhibiting tumor progression |
| Frizzled 8 | ||
| miR-100 | Downregulation | Inhibiting tumor progression |
| SFRPs | ||
| miR-27a | Downregulation | Suppressing cell growth and invasion |
| CDK8 | Downregulation | Inhibiting cell proliferation and metastasis |
| miR-107 | ||
| miR-26b |
We began by identifying 1,373 genes targeted by miRNAs that are both up-or-down-regulated, and playing a role in the Wnt pathway. These genes were chosen based on a miRDB database score of over 95. All genes with elevated scores controlled by miRNAs were carried forward for additional analysis. To map the complex protein interactions within these gene sets, we used the STRING database and Cytoscape. The resulting network from Cytoscape maps both direct (physical) and indirect (functional) relationships, combining predictive computational data, insights from cross-species studies, and data from primary databases (16).
4.2. Creating Networks and Examining Central Proteins in Protein-Protein Interactions
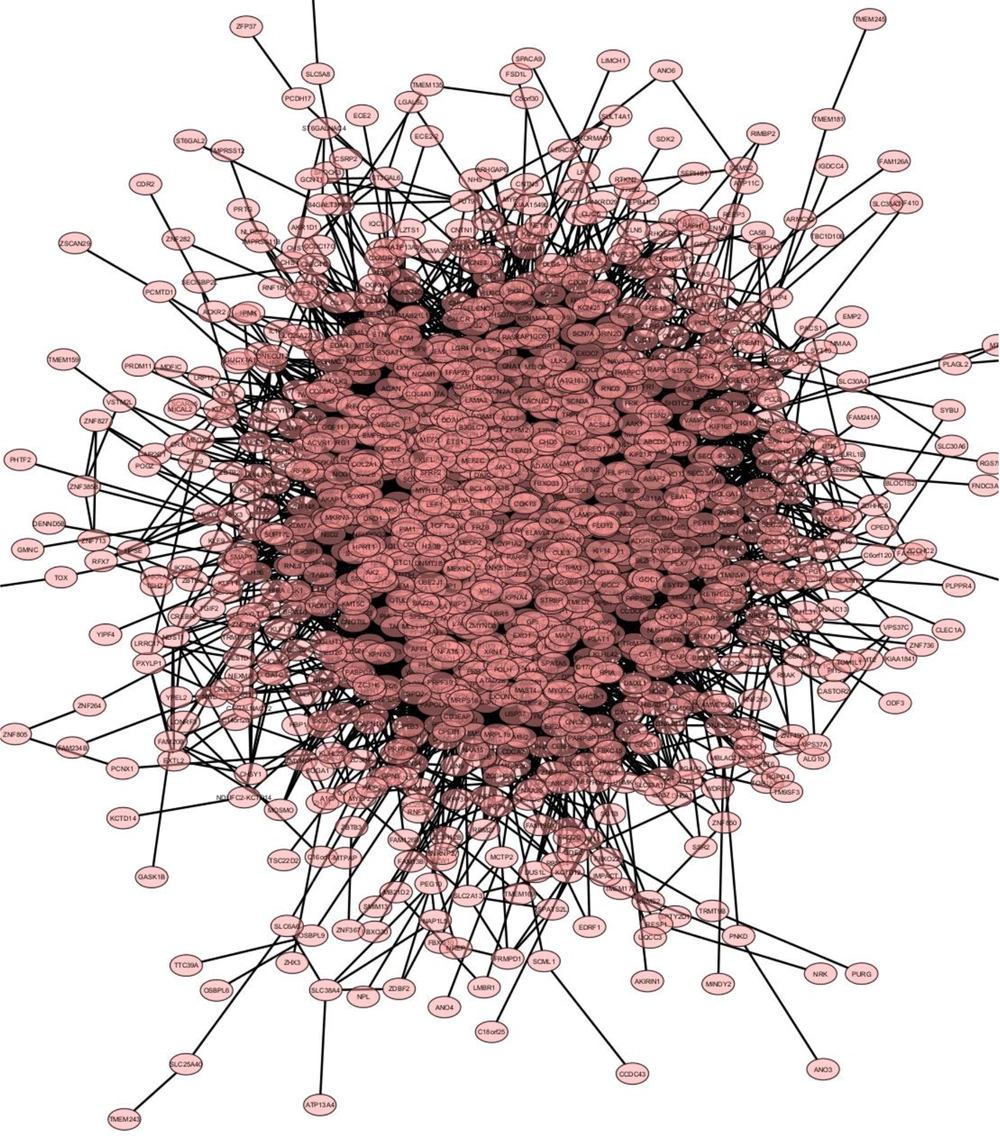
Biological networks, which depict complex biological systems, often encompass a variety of elements. Cytoscape, a powerful software tool supported by various applications, helps in the assessment and visualization of these networks. The recent StringApp 2.0, a Cytoscape application, significantly improves support for diverse networks. Figure 1 illustrates the gene network for all specified genes.
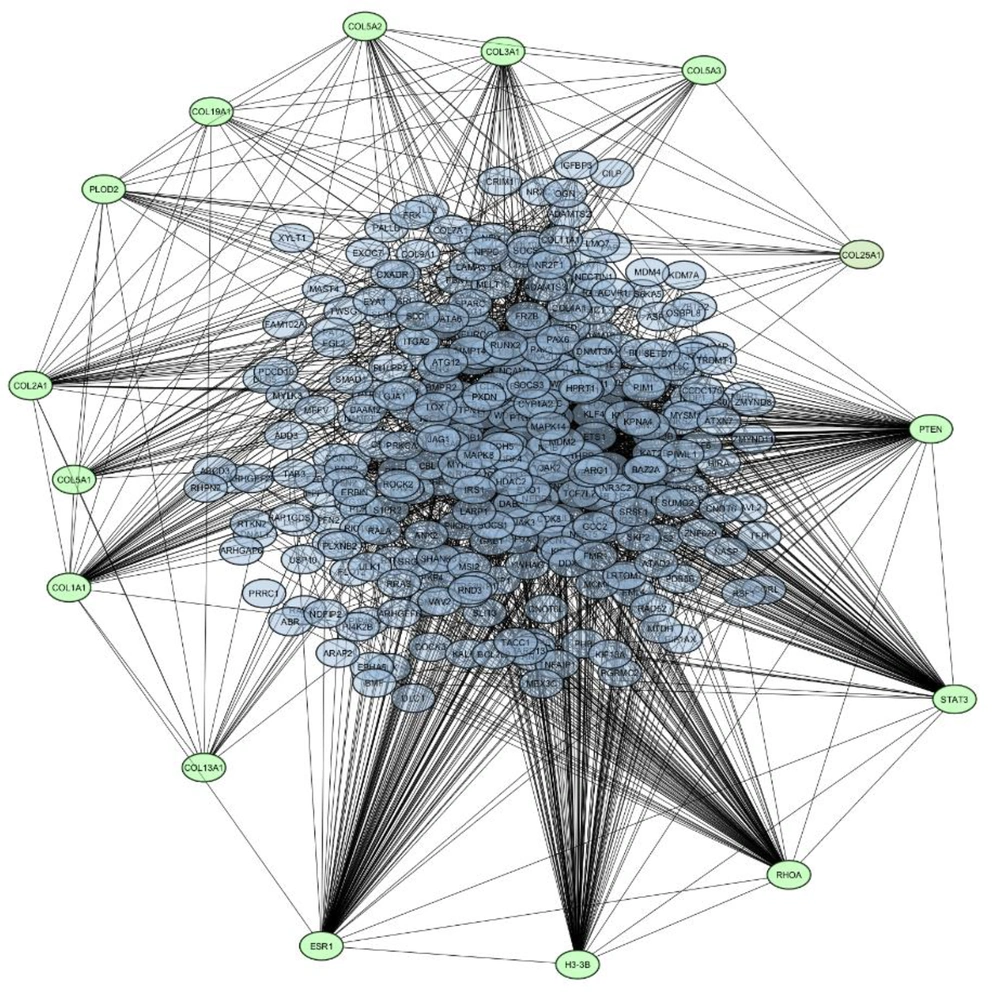
Applying four algorithms as delineated in the methods section, we have pinpointed 15 hub proteins exhibiting the highest interactions, indicating substantial roles within the network. Comprehensive descriptions of these hub proteins available in Table 2 and Figure 2 illustrate the subnetwork formed by these hubs.
| Hub Protein | Rank | Method |
|---|---|---|
| STAT3 | 1, 1 | MNC, degree |
| PTEN | 1, 1 | MNC, degree |
| ESR1 | 3, 3 | MNC, degree |
| H3-3B | 4, 4 | MNC, degree |
| RHOA | 5, 5 | MNC, degree |
| COL1A1 | 1 | MCC |
| COL3A1 | 2 | MCC |
| COL5A1 | 3 | MCC |
| COL5A2 | 4 | MCC |
| COL2A1 | 5 | MCC |
| COL25A1 | 1 | DMNC |
| COL19A1 | 2 | DMNC |
| COL13A1 | 3 | DMNC |
| PLOD2 | 4 | DMNC |
| COL5A3 | 5 | DMNC |
4.3. Functional Annotation and Pathway Enrichment Analysis Using Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Databases
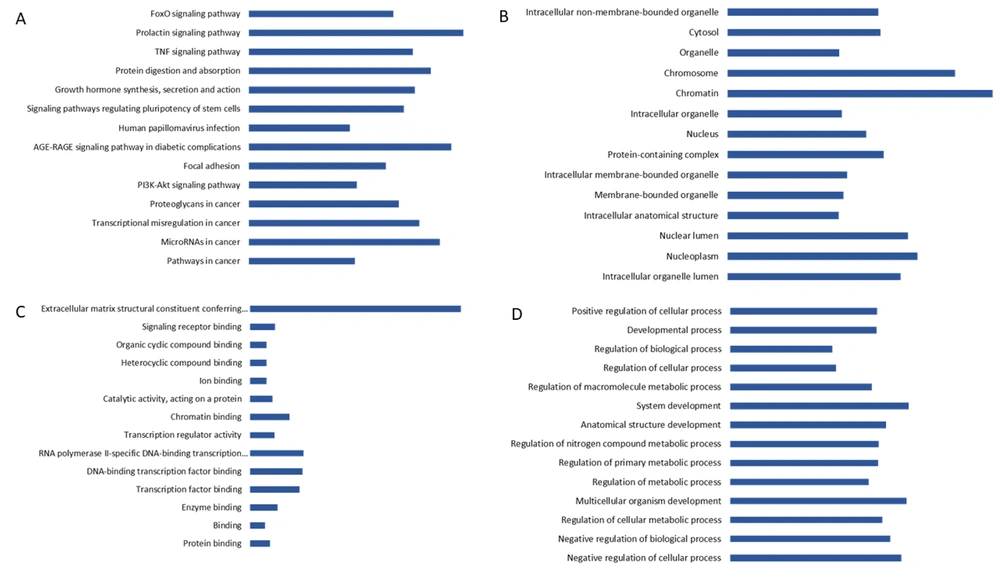
Functional annotation and pathway enrichment analyses were conducted using the GO and KEGG databases to investigate the functional relevance of the 15 identified hub proteins in breast cancer. In the GO enrichment analysis, the hub proteins were significantly involved in several BPs such as negative regulation of cellular processes, regulation of metabolic processes, and multicellular organism development. In terms of cellular component localization, the proteins were predominantly associated with the nucleoplasm, membrane-bounded organelles, and chromatin. Additionally, MF analysis revealed enrichment in protein binding, enzyme binding, and transcription factor binding, indicating their engagement in diverse intracellular interactions.
The KEGG pathway analysis further demonstrated the involvement of these hub proteins in multiple cancer-related pathways. Among the most significantly enriched pathways were “Pathways in cancer”, “MicroRNAs in cancer”, and “Transcriptional misregulation in cancer”. Furthermore, signaling cascades including the “PI3K-Akt signaling pathway” and the “TNF signaling pathway” were notably enriched, suggesting a role for these proteins in regulating cell survival, proliferation, and inflammatory signaling. Additional pathway enrichment in “Signaling pathways regulating pluripotency of stem cells” and “Protein digestion and absorption” points to their possible involvement in stemness and metabolic regulation in breast cancer cells. These findings collectively highlight the diverse biological and MFs of the hub proteins and their potential involvement in breast cancer pathogenesis (Figure 3).
4.4. Analysis of the Subnetwork Proteins by CytoCluster
The application of CytoCluster analysis allowed the identification of network clusters that hold critical implications for understanding breast cancer dynamics. CytoCluster analysis was instrumental in uncovering the organization of dysregulated miRNA-targeted genes within distinct clusters. The top 5 CytoCluster results, detailed in Table 3, provide a comprehensive view of these clusters, offering insights into their structure, size, and associated KEGG enrichment analysis.
| KEGG Enrichment Analysis | Cluster | Rank | Nodes | Edges |
|---|---|---|---|---|
| Breast cancer | 1 | 1 | 31 | 294 |
| Pathways associated with cancer | ||||
| The PI3K-Akt signaling cascade | ||||
| Prostate cancer | ||||
| Melanoma infection | ||||
| Breast cancer | 2 | 2 | 30 | 290 |
| The PI3K-Akt signaling cascade | ||||
| Pathways associated with cancer | ||||
| Melanoma | ||||
| Prostate cancer | ||||
| Breast cancer | 3 | 3 | 30 | 304 |
| Pathways associated with cancer | ||||
| The PI3K-Akt signaling cascade | ||||
| Prostate cancer | ||||
| Melanoma | ||||
| The PI3K-Akt signaling cascade | 4 | 4 | 30 | 299 |
| Breast cancer | ||||
| Pathways associated with cancer | ||||
| Resistance to EGFR tyrosine kinase inhibitors | ||||
| The role of proteoglycans in cancer development and progression | ||||
| The PI3K-Akt signaling cascade | 5 | 5 | 30 | 285 |
| Breast cancer | ||||
| Pathways associated with cancer | ||||
| Resistance to EGFR tyrosine kinase inhibitors | ||||
| The role of proteoglycans in cancer development and progression |
Abbreviation: KEGG, Kyoto Encyclopedia of Genes and Genomes.
The consistent enrichment of all clusters in the "Breast cancer" pathway signifies a direct relevance to breast cancer progression. These clusters provide a glimpse into the intricate molecular interactions within downregulated miRNA-targeted genes, potentially influencing key processes associated with breast cancer. The recurrent enrichment of the PI3K-Akt signaling pathway across clusters underscores its pivotal role in the regulatory network. Known for its involvement in cell survival and proliferation, this pathway’s prominence suggests a central influence on breast cancer-related signaling cascades. Moreover, the diversity in enriched pathways, such as "Prostate cancer", "Melanoma infection", and specific enrichments like "EGFR tyrosine kinase inhibitor resistance" and "Proteoglycans in cancer", unveils a broader impact on cancer-related processes beyond breast cancer. These findings highlight potential interplay and shared regulatory mechanisms across different cancer types.
4.5. Promoter Motif Analysis of Hub Proteins
The examination of promoter sequence patterns provides understanding of potential elements that regulate the expression of central hub proteins, unveiling their putative roles in cellular processes.
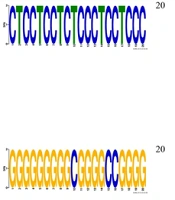
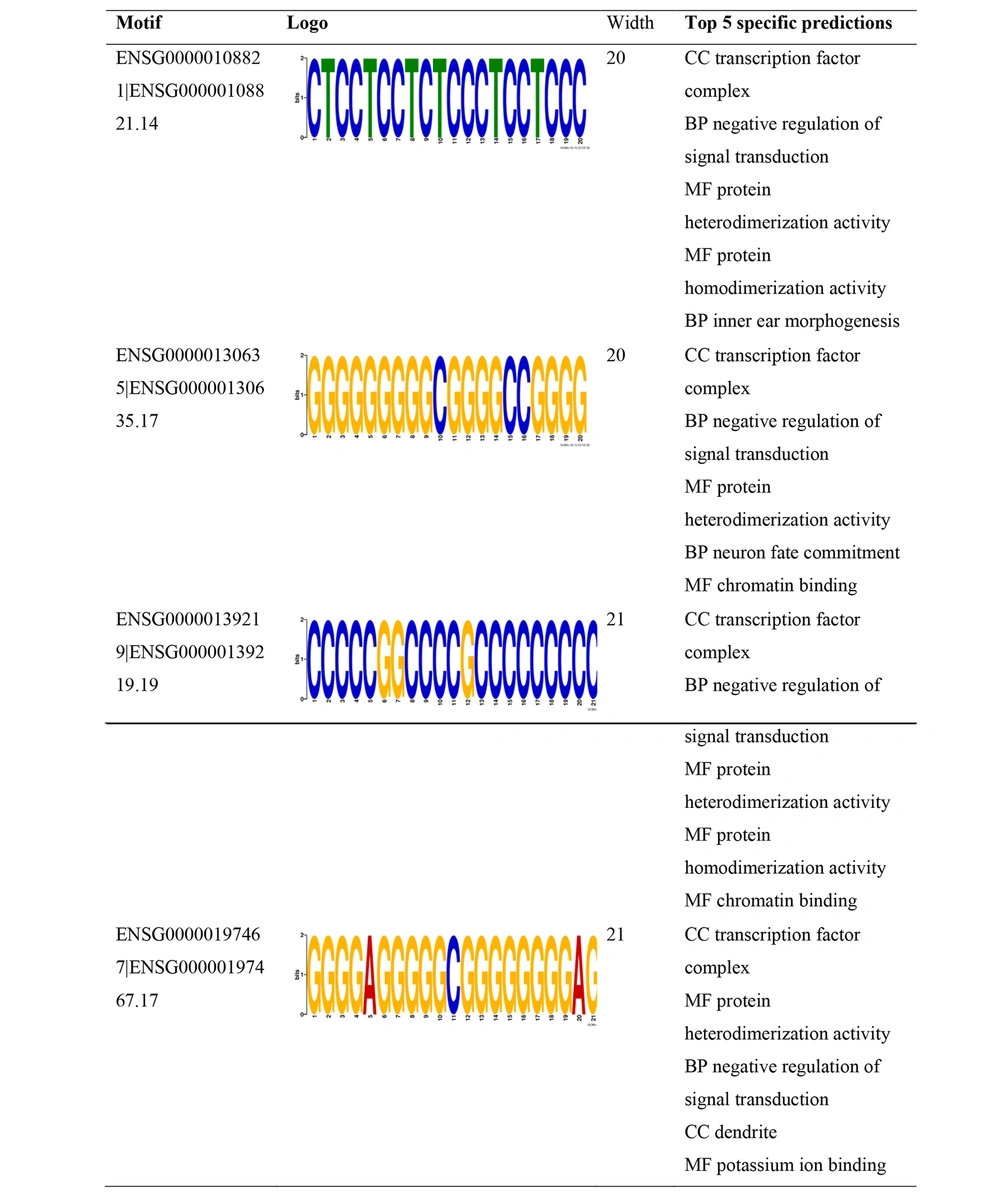
Across all hub proteins, there is a consistent motif prediction associated with the "transcription factor complex", indicating their potential involvement in transcriptional regulation. This aligns with their roles as key regulatory proteins in cellular processes. The BP predictions such as "negative regulation of signal transduction" and "inner ear morphogenesis" (in ENSG00000108821|ENSG00000108821.14) highlight the diverse functional roles of the hub proteins. These predictions suggest regulatory functions in signal transduction pathways and developmental processes. The MF predictions, including "protein heterodimerization activity" and "protein homodimerization activity", suggest the hub proteins’ involvement in forming protein complexes, potentially influencing various cellular activities. Predictions related to cellular components like "dendrite" and MF annotations like "potassium ion binding" (in ENSG00000197467|ENSG00000197467.17) hint at specific cellular localizations and ion-binding capabilities, indicating potential roles in cellular structure and ion homeostasis (Figure 4).
4.6. DrugBank Analysis of Compounds Targeting Critical Hub Proteins
The analysis conducted using DrugBank aims to identify compounds that specifically target certain hub proteins. Table 4 presents a collection of compounds that target hub proteins involved in breast cancer development. Hub proteins, including STAT3, ESR1, PTEN, and RHOA, are central to regulating essential cellular processes such as proliferation, survival, and metastasis. Drugs like acitretin (a STAT3 inhibitor) and raloxifene (an ESR1 modulator) show promising therapeutic potential by influencing these critical pathways. Natural compounds such as epigallocatechin gallate and quercetin also display inhibitory effects on cancer-related signaling. Experimental agents like alpelisib (a PI3K modulator) and collagenase Clostridium histolyticum (an enzyme that degrades collagen) further illustrate the variety of pharmacological strategies for targeting breast cancer, although no compounds were identified for the remaining hub proteins. These findings underscore the significance of hub proteins as therapeutic targets and highlight the necessity for further research to evaluate the efficacy and safety of these compounds in clinical applications.
| Drugs | Drug Group | Pharmacological Action | Type | Actions |
|---|---|---|---|---|
| Acitretin | Approved, investigational | Unknown | STAT3 | Inhibitor |
| Atiprimod | Investigational | Unknown | Inhibitor | |
| Celecoxib | Approved, investigational | Unknown | Inhibitor | |
| Diosmetin | Approved | Unknown | Inhibitor | |
| ENMD-1198 | Experimental | Unknown | ||
| Epigallocatechin gallate | Investigational | Unknown | Inhibitor | |
| Golotimod | Investigational | Unknown | Inhibitor | |
| Luteolin | Investigational | Unknown | Inhibitor | |
| MOL-4239 | Investigational | Unknown | Inhibitor | |
| Napabucasin | Investigational | Unknown | Inhibitor | |
| NT-219 | Investigational | Unknown | Inhibitor | |
| OPB-111077 | Investigational | Unknown | Inhibitor | |
| Quercetin | Investigational | Unknown | Inhibitor | |
| TTI-101 | Investigational | Unknown | Inhibitor | |
| WP-1066 | Investigational | Unknown | Inhibitor | |
| Phosphatidylethanolamine | Experimental | Unknown | PTEN | Substrate |
| Benzophenone | Approved | Unknown | ESR1 | |
| Enzacamene | Approved | Unknown | ||
| Estrone | Approved | Yes | Agonist | |
| Estrone sulfate | Approved | Yes | Agonist | |
| Ethynodiol diacetate | Approved | Yes | Agonist | |
| Homosalate | Approved | Unknown | ||
| Nomegestrol acetate | Approved | Unknown | Modulator | |
| Norethynodrel | Approved | Unknown | ||
| Octocrylene | Approved | Unknown | ||
| Ospemifene | Approved | Yes | Antagonistagonist | |
| Oxybenzone | Approved | No | ||
| Polyestradiol phosphate | Approved | Yes | Agonist | |
| Quinestrol | Approved | Yes | Agonistmodulator | |
| Raloxifene | Approved | Yes | Agonist | |
| Synthetic conjugated estrogens, A | Approved | Yes | Agonist | |
| Synthetic conjugated estrogens, B | Approved | Yes | Agonist | |
| Zinc chloride | Approved | Unknown | Binder | |
| Zinc sulfate, unspecified form | Approved | Unknown | Binder | |
| Fluoxymesterone | Approved, illicit | Yes | Antagonist | |
| Alpelisib | Approved, investigational | Yes | Modulator | |
| Bazedoxifene | Approved, investigational | Yes | Antagonistagonist | |
| Guanosine-5'-diphosphate | Experimental | Unknown | RHOA | Guanosine-5'-Diphosphate |
| Clove oil | Approved, nutraceutical | Unknown | COL1A1 | |
| Collagenase clostridium histolyticum | Approved, investigational | Yes | Cleavage | |
| Halofuginone | Investigational, vet-approved | Unknown | ||
| PR-15 | Investigational | Unknown | ||
| Von Willebrand factor human | Approved, investigational | Yes | Binder | |
| Vonicog alfa | Approved, investigational | Yes | Binder | |
| Collagenase clostridium histolyticum | Approved, investigational | Yes | COL3A1 | Cleavage |
| PR-15 | Investigational | Unknown | ||
| Collagenase clostridium histolyticum | Approved, investigational | Yes | COL2A1 | Cleavage |
| Ascorbic acid | Approved, investigational, nutraceutical | Unknown | PLOD2 | Cofactor |
5. Discussion
Numerous studies underscore the potential of miRNAs serving as indicators for cancer detection, outcome prediction, and treatment strategies intervention, highlighting the importance of additional research and confirmation. The MiRNA aberrant regulation in human cancers, driven by processes including miRNA gene reinforcement or gene loss, irregular gene expression patterns, epigenetic alterations, and disruptions in miRNA biogenesis, contributes significantly to tumor initiation, progression, metastasis, and resistance to therapy (14). The miRNAs function as both cancer-promoting genes and tumor-inhibiting genes, influencing key carcinoma characteristics, encompassing uncontrolled cell proliferation, evasion of growth suppression, resistance to apoptosis, activation of invasion and metastasis, and stimulation of angiogenesis. Using computational resources, our research investigates the complex interplay between miRNAs that regulate the Wnt/β-catenin cellular signaling cascade and their role in breast carcinoma initiation and advancement. These miRNAs are critical due to their activation and interaction with other essential cellular pathways (15, 16).
The PPI networks provide a mathematical representation of physical interactions between proteins in cellular environments. These interactions are highly specific, taking place in well-defined binding domains, and hold significant biological relevance by fulfilling distinct functional roles. In recent years, scientific focus has shifted toward analyzing protein substructures, employing iterative interaction algorithms to predict protein interactions and elucidate their functional roles. This methodology allows for the scrutiny of critical proteins vital for optimal targeting. In Figure 1, proteins directly associated with aberrantly miRNAs play crucial contributions in fundamental signaling cascades involved in cellular growth and division in subsequent sections, our objective is to offer a more detailed classification and comprehension of these proteins, pinpointing those with the highest number of interactions (17, 18).
RHOA is involved in cell migration and invasion. Dysregulation of RHOA has been linked to enhanced metastatic potential in cells of breast cancer, indicating a potential role in the spread of cancer. COL5A3 is a collagen gene (19). Altered collagen expression is associated with changes in the tumor microenvironment, impacting cancer cell behavior, including migration and invasion (20).
Again COL19A1, COL1A1, COL5A1, COL2A1, COL3A1, COL25A1, and COL13A1 role in the extracellular matrix suggests a potential impact on tissue remodeling, which could influence the advancement and invasion cells of breast cancer (21). ESR1 encodes estrogen receptor alpha protein. Mutations or alterations in ESR1 are crucial in hormone receptor-positive breast cancers, influencing treatment response to hormone therapies (22). H3F3B encodes a histone variant. Alterations in histones can affect chromatin structure, potentially influencing gene expression and contributing to breast cancer development (23). PLOD2 is associated with collagen modification. Changes in collagen stability may influence the extracellular matrix in breast cancer and play a role in tumor advancement (24). STAT3 is a signaling protein involved in cellular viability and proliferation. Its activation has been associated with breast cancer development and progression, positioning it as a promising candidate for therapeutic intervention (25). PTEN acts as a tumor suppressor gene. Absence of PTEN activity, often due to genetic alterations, is linked to breast cancer development, as PTEN regulates cell growth and division (26).
The functional enrichment results suggest that the identified hub proteins play critical roles in several key mechanisms associated with breast cancer development and progression. Their involvement in the regulation of cellular and metabolic processes, as well as multicellular development, reflects their potential impact on fundamental aspects of tumor biology, including proliferation, differentiation, and survival. Localization to the nucleoplasm and chromatin emphasizes their possible functions in gene expression regulation and epigenetic control, which are frequently altered in cancer cells. Moreover, the diversity of MFs, particularly their capacity for protein and transcription factor binding, highlights their regulatory complexity and potential to influence multiple signaling axes. The enrichment in PI3K-Akt and TNF signaling pathways supports their likely involvement in oncogenic signaling that governs cell viability, immune responses, and tumor microenvironment interactions. Notably, their association with stem cell-related pathways underscores a potential link to cancer stemness, a feature associated with tumor initiation, resistance to therapy, and recurrence. The unexpected connection to protein digestion and absorption pathways may also reflect an adaptive metabolic reprogramming in cancer cells, enabling efficient nutrient uptake to sustain rapid growth. Overall, these results provide valuable insights into the molecular mechanisms driven by hub proteins and reinforce their relevance as potential biomarkers or therapeutic targets in breast cancer (Figure 3).
CytoCluster provides a glimpse into the intricate molecular interactions within downregulated miRNA-targeted genes, potentially influencing key processes associated with breast cancer. The prominence of the PI3K-Akt pathway suggests a central influence on breast cancer-related signaling cascades, known for its involvement in cell survival and proliferation. The enrichment of pathways beyond breast cancer implies a broader regulatory network, potentially shared across multiple cancers. These findings highlight potential interplay and shared mechanisms between breast cancer and other malignancies, offering new perspectives for future research. In conclusion, the results of the CytoCluster analysis offer a nuanced understanding of the network organization of downregulated miRNA-targeted genes, emphasizing their direct relevance to breast cancer and providing valuable insights into potential therapeutic targets and mechanisms of cancer progression. Further experimental validations and functional studies are warranted to unravel the specific roles of these clusters and their implications for targeted therapeutic interventions in breast cancer and potentially other cancer types (Table 3).
Predicted motifs and functional annotations indicate that hub proteins may play key regulatory roles in diverse cellular processes, particularly in signal transduction and development. The consistent identification of transcription factor-related motifs highlights their potential involvement in transcriptional control mechanisms. Further insights from MF predictions, such as dimerization activities and ion-binding capabilities, suggest the formation of protein complexes and participation in cellular signaling and homeostasis. Localization patterns, including associations with dendritic structures, reinforce their functional importance in specialized cellular compartments. Overall, these findings broaden the understanding of the regulatory framework influencing hub protein expression and function. The results provide a compelling basis for future experimental validation and investigation into the biological relevance of these computational predictions (Figure 4).
The identification of approved, investigational, and experimental compounds targeting hub proteins such as STAT3, ESR1, PTEN, and RHOA reinforces the therapeutic relevance of these molecules in breast cancer management. The presence of both synthetic agents (e.g., acitretin, raloxifene, alpelisib) and natural compounds (e.g., epigallocatechin gallate, quercetin) highlights the diversity of pharmacological strategies aimed at modulating critical signaling pathways. Notably, the absence of drug associations for several hub proteins underscores a need for continued drug discovery efforts. These insights emphasize the translational potential of network-based drug screening and the importance of validating predicted interactions in preclinical and clinical studies (Table 4).
5.1. Conclusions
Our integrative systems biology analysis delves into the complexities of miRNA regulation in the Wnt/β-catenin pathway, unraveling its critical role in breast cancer. Through meticulous exploration of dysregulated miRNA expression, PPIs, hub protein identification, and promoter motif analysis, we unveiled a comprehensive understanding of the molecular landscape. The study identified key miRNAs involved in modulating the Wnt signaling cascade in breast neoplasm and elucidated their functional significance through GO and KEGG analyses. CytoCluster analysis further emphasized the direct relevance of dysregulated miRNA-targeted genes to breast cancer, while promoter motif analysis shed light on potential regulatory components controlling the expression of central hub proteins. Collectively, these findings contribute to a nuanced comprehension of breast cancer pathogenesis, providing perspectives on potential targets for therapeutic intervention and signaling cascades crucial for personalized treatment strategies. Future experimental validations are imperative to translate these discoveries into actionable interventions for advancing breast cancer diagnosis, prognosis, and treatment modalities.