1. Background
Colorectal carcinoma (CRC) is the third most malignant tumor in the world (1) with approximately 10 percent of all cancers incidence and mortality (2). It has been shown that CRC will be a preventable malignancy when precancerous lesions are diagnosed early. Despite considerable progress that has been made over the past two decades in CRC diagnostic procedures and therapeutic strategies (3-5), however still many of the CRC patients, especially those who are diagnosed in advanced stages; do not suitably respond to treatment. Thus intensive researches should be done to improve our understanding of biology, genetics and epigenetic alterations in CRC for identifying diagnostic, prognostic and predictive biomarkers and finally more effective therapeutic procedures. Recent studies have shown that expression of small RNA molecules named microRNA is significantly associated with formation and progression of CRC (6-8). MicroRNAs (miRNAs) are a class of small, non-coding, single-stranded, endogenous and highly conserved RNA molecules which post-transcriptionally modulate the expression of their target genes (9). MiRNAs can influence on colorectal carcinogenesis by direct regulation of their target mRNAs as oncogenes or tumor suppressor genes. A significant advantage of miRNAs as useful molecular tools comes from the fact that a single miRNA can modulate the expression of several mRNAs. So, for identifying the markers with greater specificities and sensitivities, working on miRNA is preferred (10). Another advantage of using the miRNA is their high stability in different physical situations and resistance to storage and handling in different biological samples (11, 12). Previous studies demonstrated that miRNAs are highly stable in serum or plasma samples with appropriate concentration (13-15). Valadi et al. in 2007 reported the secretion of miRNAs in circulation system through exosomal compartments (16). The tumor-specific expression profile of miRNAs in the body fluids as well as tumor tissues is a suitable biomarker for a variety of cancers (17-19). Moreover body fluids provide constant, non-invasive source of miRNAs as cancer biomarkers.
2. Objectives
In the present study we have compared the miRNAs expression profiles in plasma of CRC patients and healthy individuals hoping to find the miRNAs with significant altered expression in the patient group compared to the control group.
3. Materials and Methods
3.1. Subjects and Blood Samples
In this study, pre-operative plasma from 61 recently diagnosed colorectal cancer patients (including 13 stage I, 41 stage II and 7 stage III patients) and 24 (age and gender-matched) healthy subjects as the control were collected between 2012 and 2013 at Digestive Disease Research Institute (DDRI), Shariati Hospital, Tehran, Iran (Table 1). Tumors were staged according to the tumor–node–metastasis (TNM) staging system guidelines. No significant differences were observed between the CRC patients and controls in the distribution of gender (P value = 0.512, chi-square test) and age (P value = 0.8, Independent Samples t test). Normal subjects were asymptomatic individuals recruited from a colonoscopy screening. The CRC patients group consisted of 27 women and 34 men with a median age of 66 years (range, 45 - 80) and the healthy subjects consisted of 10 women and 14 men with a median age of 63 years (range, 47 - 77). The patients with a history of familial adenomatous polyposis or hereditary non-polyposis CRC or previous history of malignant tumors were excluded from this study. An amount of 5 to 10 milliliters of whole blood were obtained from each participant. The plasma was obtained by centrifugation at 1200 gr for 10 minutes at 4°C. The supernatant plasma was quickly frozen in -70°C until analysis. Informed consents were obtained from all participants for the use of their blood samples in this study. This study was approved by the Clinical Research Ethics Committee of Shariati Hospital, Tehran University of Medical Sciences.
| 64.13 ± 8.673 | 61.96 ± 8.67 | |
| Male | 34 | 14 |
| Female | 27 | 10 |
| I | 13 | - |
| II | 41 | - |
| III | 7 | - |
| Colon | 52 | - |
| Rectum | 9 | - |
Patient Characteristics for Plasma miRNA Analysis
3.2. RNA Isolation
Total RNA, including miRNA, was extracted from plasma samples by using the miRNA easy Serum/Plasma Kit (Qiagen) following the manufacturer’s instruction. For normalization of sample-to-sample variation during RNA isolation and as internal control, a synthetic miRNA (cel-miR-39 from C. elegans) was added into each denatured sample in equal amounts. RNA concentration and integrity were quantified using NanoDrop-1000 Spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE) and Agilent’s Bioanalyzer (Agilent Technologies, Palo Alto, CA).
3.3. MiRNA Microarray Expression Profiling and Data Analysis
The miRNA expression profiles were generated by using the Agilent's miRNA Microarray system (V19), (Agilent Technologies, Santa Clara, CA), on plasma samples from 37 CRCs (including 9 stage I, 25 stage II and 3 stage III patients) and 8 normal controls. This array detects 2006 human mature miRNAs. Briefly, in labeling step 100 ng of the diluted total RNA was dephosphorylate by calf intestinal alkaline phosphatase followed by denaturation with 2.8 µL of 100% DMSO and ligation to Cyanine3-pCp using T4 RNA Ligase. After drying the samples using a vacuum concentrator and treatment with GE blocking agent and Hi-RPM hybridization buffer, samples were loaded on Agilent SureHyb chamber. In hybridization step, samples were hybridized overnight to miRNA bead chip in a hybridization oven at 55°C. After washing the samples were scanned by Agilent's Microarray Scanner. Microarray data processing was generated in the agilent feature extraction software and used for further analysis with GeneSpring GX Analysis software (Agilent Technologies Inc., USA).
3.4. Quantification of MiRNAs by qRT-PCR
100 ng of extracted total RNA was reverse transcribed with a miScript Reverse Transcription Kit (Qiagen, Valencia, CA), in a 20 μL reaction volume according to the manufacture's protocol. MiRNA expression levels were quantified in duplicate by Real-time PCR using SYBR Green miScript real-time PCR system (Qiagen, Valencia, CA) according to the manufacturer's instructions on LightCycler v.3.5 system (Roche Applied Science, Mannheim, Germany). Product amplification was performed using miRNA-specific primers and universal primers (Qiagen, Valencia, CA). In brief, the 20 μL PCR reaction mixture included 2 μL of RT product (10 ng), 10 μL 2x QuantiTect SYBR Green PCR Master Mix, PCR primer and universal primer each one 2 μL and 4 μL nuclease free water, was incubated at 95°C for 15 minutes., followed by 50 cycles of 94°C for 15 seconds, 55°C for 20 seconds and 72°C for 20 seconds. The expression levels of each miRNA were normalized against miR-16 expression and the threshold cycle (Ct) values of 40 or greater were defined as undetectable. The relative expression of plasma miRNA levels between two groups was calculated using the 2-∆∆Ct method. GraphPad Prism 6 was used for data analysis and preparing the graphs.
4. Results
4.1. MicroRNA Expression Profiling
The extracted total RNA concentration ranges were from 21.1 to 54.87 ng/μL in plasma samples of CRC patients and normal group. By data analysis of miRNA microarray on 37 CRC patients and 8 normal subjects, using precise significance criteria of a two-fold or greater difference in expression level and P value < 0.05, in GeneSpring GX software, we found that six miRNAs (miR-142-3p, miR-26a-5p, miR-27a-3p, miR-326, miR-331-3p and miR-6073) were significantly downregulated in CRC patients compared to the healthy subjects. The list of these differentially expressed miRNAs is provided in Table 2.
| miR Name | MiRNAs With Low Expression in Plasma of CRC Patients | |
|---|---|---|
| P Value | Fold Change | |
| 0.029 | 11.31 | |
| 0.023 | 10.97 | |
| 0.046 | 9.58 | |
| 0.038 | 5.02 | |
| 0.033 | 9.29 | |
| 0.048 | 10.17 | |
The Most Differentially Expressed MiRNAs in CRC Patients Versus Normal Subjects
4.2. Validation Study by qRT-PCR
Among the six differentially expressed miRNAs in our profiling study, miR-142-3p and miR-26a-5p were chosen for further qRT-PCR validation study, because they showed the most changes in expression level in miRNA microarray step. In addition, their misexpressions have been shown in different types of cancers. The expression levels of miR-142-3p and miR-26a-5p were evaluated in the plasma from 61 CRC patients and 24 healthy subjects (including the samples used for miRNA microarray analysis).
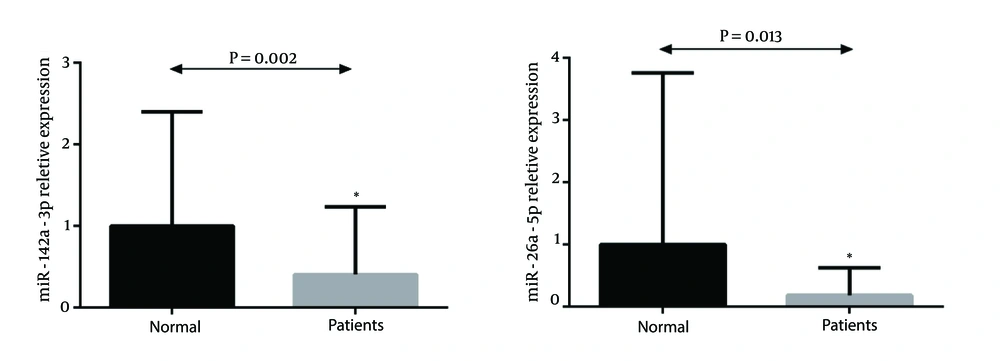
Real-time data analysis showed significant downregulation of miR-142-3p and miR-26a-5p in CRC patients compared to the normal subjects (P value = 0.002 and 0.013 respectively) (Figure 1 A and B).
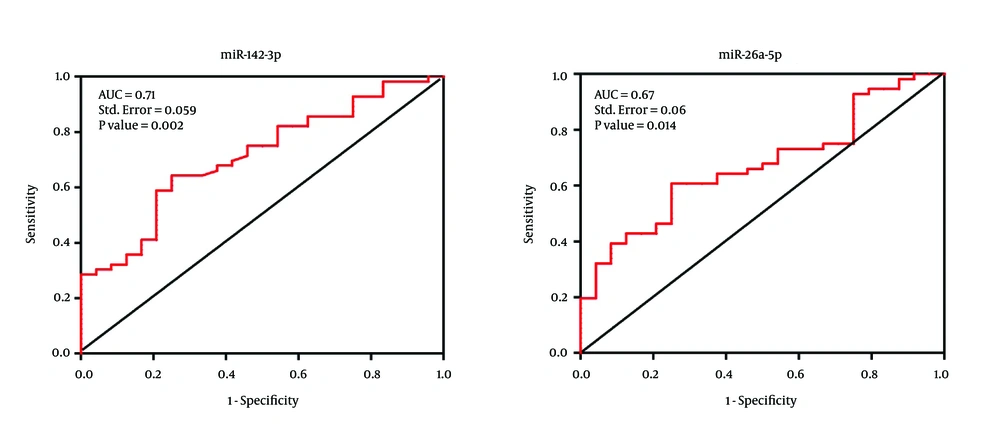
In order to evaluate the diagnostic value of the miR-142-3p and mir-26a-5p in CRC patients, ROC curve analysis was performed. These data revealed that miR-142-3p can potentially be used as a diagnostic biomarker for distinguishing between CRC patients and normal individuals. The area under the curve (AUC value) for miR-142-3p was 71% with the standard error of 0.059 (P value = 0.002) while the AUC value for miR-26a-5p was 67% with the standard error of 0.06 (P value = 0.014) (Figure 2).
5. Discussion
Previous studies have shown that compared to the mRNA levels, miRNA analysis may be more efficient in understanding the biology of the disease (20, 21). The main objective of this study was to identify altered expression circulating miRNAs in patients with colorectal carcinoma compared to healthy participants. Circulating miRNAs are reflective of alterations in tissue expression of miRNAs and are emerging as biomarkers in many malignancies, including colorectal cancer.
In the present study, to identify differentially expressed circulating miRNAs we have firstly compared the expression profile of miRNAs in the plasma samples from CRC patients compared to healthy controls using a miRNA microarray method and six miRNAs (miR-142-3p, miR-26a-5p, miR-27a-3p, miR-326, miR-331-3p and miR-6073) were found significantly downregulated in CRC patients. In order to validate our data, expression levels of miR-142-3p and miR-26a were evaluated in a subsequent validation study using qRT-PCR method and the results showed that the plasma levels of these two selected miRNAs were significantly decreased in CRC patients compared to the normal group.
Another downregulated miRNA, miR-326 has been already found to be downregulated in advanced breast cancer (22) and also in glioblastoma (23). Furthermore miR-326 has been identified as a potential marker associated with osseous metastasis in a lung cancer (24). MiR-331-3p expression has demonstrated to be decreased in some malignancy including breast, prostate, and pancreas cancer via targeting of human epidermal growth factor receptor 2 (HER2) (25, 26) and in glioblastoma through targeting the neuropilin-2 (NRP-2) (27). Overexpression of the long non-coding RNA HOTAIR (Hox transcript antisense intergenic RNA) has been identified in a range of tumors, including those of the colorectal cancer, hepatocellular carcinoma, pancreatic and gastric cancer, and have been shown that miR-331-3p can directly bind to HOTAIR (28). In a recent study by Wei Shen et al. miR-142-3p was found to be downregulated in colon cancer (29). They reported that reduced levels of miR-142-3p as a tumor suppressor is associated with upregulation of CD133, ATP binding cassette G2 (ABCG2), and leucine-rich-repeat-containing G-protein-coupled receptor 5 (Lgr5) by binding to their 3’-UTRs and coding sequences. Upregulation of these genes are associated with poor prognosis in colon cancer. In addition Zhou et al. found that miR-142-3p may be involved in the regulation of cell proliferation in colorectal cancer through targeting the transcription factor 7 (TCF7) (30). Another previous study revealed that miR-142-3p could regulate FASN expression which is involved in cancer cells proliferation and metastasis in CRC (31). Previously has been reported that miR-26a could functions as a potential tumor-suppressor by targeting the MYC oncogene (32, 33). Furthermore low expression levels of miR-26a has been shown in Melanoma (34), lung cancer (35), breast cancer (36) and nasopharyngeal carcinoma (33). Most recently, Jinushi et al., identified that miR-26a expression level may be used as a prognostic biomarker in CRC (37).
This research will serve as a base for future studies. Taken together our finding indicated that the differential expression of circulating miRNAs may have potential value in CRC diagnosis and might serve as novel minimally invasive molecular biomarker for this malignancy. Although a limitation of this study was the relatively small number of patients and controls and further large-scale experimental investigations are needed to verify the results of this study.