1. Background
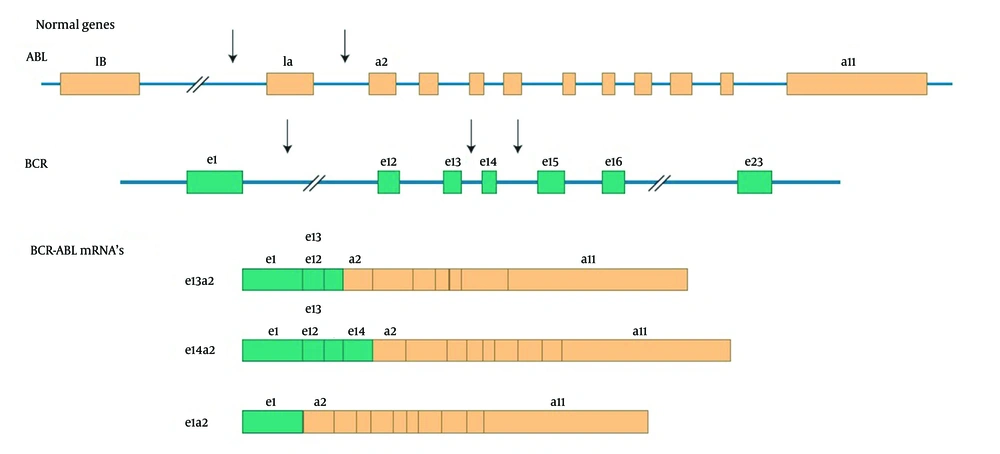
Chronic myelogenous leukemia (CML) is a hematopoietic stem cell malignancy accounted for 15% to 20% of diagnosed adult CML patients (1). The hallmark of CML in 95% of patients is a chromosomal reciprocal translocation which forms a kind of BCR-ABL fusion gene and an abnormal short chromosome 22, defined as the Philadelphia chromosome (1, 2). This abnormal chromosome can be detected in mitotic cells under microscope using traditional cytogenetic techniques to prepare karyotype for the cases or using the fluorescent in situ hybridization (FISH) technique in which probes hybridize to the fusion gene and radiate fluorescent signals. BCR gene and ABL proto-oncogene are located on chromosomes 22 and 9, respectively. The break point junctions in BCR-ABL mostly occur in exon b2 or b3 of BCR gene and exon a2 of ABL gene to create b2a2 or b3a2 fusion (Figure 1). Transcripts of the fusion gene are mostly p190 and p210. The presence of constitutively activated tyrosine kinase BCR-ABL has a main role in the pathogenesis of disease (2). The discussed leukemia starts with indolent chronic phase (CP) with variable durations that would progress to the second stage of disease, accelerated phase (AP), if left untreated and within a short time blastic phase (BP) is occurred (3).
The transcripts of the BCR-ABL fusion gene usually contain one of the two BCR-ABL junctions: e13a2 (b2a2) and e14a2 (b3a2) (4).
2. Objectives
Treatments of CML demonstrate a model for targeted cancer therapy. Acceptable results obtained from tyrosine kinase inhibitor treatment in struggling with cancer cells, changed the latest process of CML patient’s treatment with interferon alpha and busulfan (5). Tyrosine kinase inhibitors represent a competition with ATP to block BCR-ABL kinase activity and progression of disease (6).
Imatinib mesylate was established in 2001 as the first line and golden standard treatment for Ph + CML patients and is known as the selective inhibitor of BCR-ABL tyrosine kinase (3). Imatinib binds to BCR-ABL at ATP binding site, so the tyrosine substrate cannot be phosphorylated and do not consequently interact with the tyrosine kinase protein, which promotes white blood cells. Imatinib is associated with a complete cytogenetic response (CcyR) rate of 87%. More than 90% of cases show complete hematologic response (CHR). A progression rate to AP or BP is assessed in 7% of cases. It is estimated that 89% of recently diagnosed CML patients show five years survival under Imatinib therapy (7). Although Imatinib is the golden standard of CML treatment and induces high rates of hematologic and cytogenetic responses in Ph + CML patients, 15% of patients reveal resistance to Imatinib and so less affirmative outcomes are achieved (5). Primary and acquired resistance can be seen in CML patients who are under Imatinib therapy (7). In primary resistance, no initial landmark response can be defined, while Imatinib acquired resistance is defined as failure to attain CHR within three months of initiation therapy, or any cytogenetic response (CyR) within six months or major cytogenetic response (McyR) (Ph + < 35%) within 12 months or evolvement of cytogenetic or hematologic relapse (8). The third popular response is molecular response. If at least 1 log increases, according to international scale (IS) in BCR-ABL/ABL ratio was seen, failure to achieve molecular response is believed. However, escalation of BCR-ABL/ABL level up to at least five folds from the baseline, which is achieved up to that time, was considered as molecular failure (5). It is reported that development of resistance against Imatinib in CML patients is defined with a range of different mechanisms. Two important mechanisms of this resistance range are:
1) BCR-ABL dependent and 2) BCR-ABL independent pathways.
BCR-ABL dependent mechanisms are the most significant cause of Imatinib resistance assorted with genomic amplification, BCR-ABL fusion gene over expression and point mutations in tyrosine kinase domain (7). To that extent, more than hundred mutations have been identified in tyrosine kinase domain of BCR-ABL fusion gene (7). These mutations have been detected in 40% to 60% of Imatinib resistant patients (5). In spite of a broad range of studies reporting different mutations with various frequencies in tyrosine kinase domain, no reports are accessible from Iranian population about the correlation of BCR-ABL tyrosine kinase domain mutations and Imatinib resistance. Resistance to Imatinib may also be due to formation of structural cytogenetic abnormalities, which leads to BCR-ABL independent proliferation of leukemic cells and progression of disease (9). In previous studies, a wide range of ABL kinase domain mutations with different frequencies were reported. However, no data was found on Iranian CML resistant patients. In this study, we investigated the frequency of BCR-ABL tyrosine kinase domain mutations on 39 Iranian CML Imatinib resistant patients using RFLP method and confirming the results by applying direct sequencing. This study tried to brighten the hypothesis that the occurrence of mutations would increase the BCR-ABL level and whether this increase can be used as a sensitive criteria for screening ABL kinase domain mutations (1).
3. Patients and Methods
3.1. Patients
In this study, we investigated the frequency of BCR-ABL tyrosine kinase domain mutations in 39 CML patients with Imatinib resistance from 135 CML patients, who referred to Payvand Clinical and Specialty Laboratory. Baseline assessment for ABL mutations status was available for 39 of these 135 patients between May 2013 and June 2014.
3.2. Inclusion Criteria
Patient inclusion was according to the criteria defined by the European Leukemia Net in 2013 (10). To consider patients as resistant to Imatinib, firstly, they had to be under Imatinib therapy with a dose of at least 400 mg daily. Secondly, they had to have failed to achieve CHR after three months of therapy in which white blood cells (WBC) count was more than 10 × 109/L or platelet count was more than 450 × 109/L. In addition, peripheral blasts had to be seen in the blood smear and also immature granulocytes had to be more than 5%. Moreover, patients with transformation to accelerated phase (AP) or blastic phase (BP) after a period of time remaining at chronic phase (CP), were characterized as Imatinib resistant. Furthermore, patients who had been under tyrosine kinase inhibitors (TKIs) therapy for more than four years, had used second or third generation of TKI drugs or undergone Imatinib dose escalations from 400 mg up to 500 mg or 600 mg daily, entered the study. This study was reviewed and approved by Tehran Azad University Ethics Committee. Prior initiation of study, all candidates were provided a voluntary written informed consent.
3.3. BCR-ABL Mutation Analysis
Peripheral blood was collected in EDTA vacuum tubes. Total RNA was extracted from the buffy coat using QIA amp RNA blood mini extraction kit (QIA-GEN, Germany) according to the manufacture’s protocol. Following to that, c-DNA synthesis using c-DNA synthesis kit (Bioline, UK) was performed. BCR-ABL mutations analysis was performed using restriction fragment length polymorphism (RFLP) method for four most common (Y253H, T315I, E255K and M351T) reported mutations. The results were confirmed by direct sequencing (9).
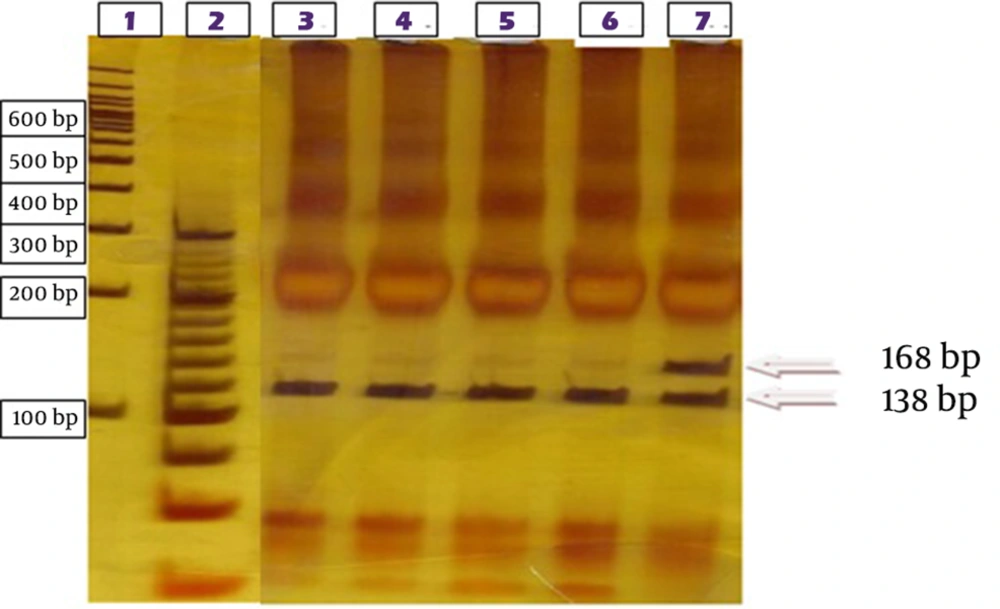
Real-time quantitative polymerase chain reaction was performed (Jena BioScience, qPCR Probes Master, Germany) to determine the copy numbers of BCR-ABL transcripts and ABL as the control gene. At the first step, PCR was performed to amplify BCR exon13 (ACAGCATTCCGCTGACCATCAATAAG) junked to ABL exon 7 (AGACGTCGGACTTGATGGAGAACT) in CML patients. The PCR product length, seen by gel electrophoresis on 1.8% ethidium bromide-stained agarose gel must be around 675 bp. The product contained different mutation sites encoding ABL amino acids 207 to 414. At the second step, specific PCR was performed for common mutations (Y253H, T315I, and E255K and M351T). Mutation specific restriction enzymes (RsaI, DdeI, MnlI and NcoI) were applied for unveiling the proportion of mutated and unmutated alleles (11). In the present study, the occurrence of mutations caused loss of restriction sites in ABL tyrosine kinase domain. A difference between mutated and unmutated digestion products was observed using gel electrophoresis on 6% silver-stained acrylamide gel (Table 1).
| Mutation | Forward Primers | Reverse Primers | Restriction Enzyme | Size of Digested Unmutated Fragment, bp | Size of Undigested Mutant Fragment, bp |
|---|---|---|---|---|---|
| TCACCACGCTCCATTATCCA | TCTTCCACACGCCCTCaTAaACCT | RsaI | 138 + 30 | 168 | |
| TCACCACGCTCCATTATCCA | CTTCCACACGCtCTCGTACA | MnlI | 136 + 31 | 167 | |
| AAGACCTTGAAGGAGGACACCATG | GTTGCACTCCaTCAaGTAGTCCA | DdeI | 134 + 49 | 183 | |
| CCCGTTCTATATCATCACTGAGTTC | AGACGTCGGACTTGATGGAGAACT | NcoI | 218 + 123 | 341 |
Mutation specific restriction digestion sites led to loss of RsaI for Y253H mutation detection, MnlI for E255K, DdeI for T315I and NcoI for M351T. Electrophoresis on polyacrylamide gel was performed and silver-nitrate used to stain acrylamide gel. For digestion results approval, direct sequencing was performed. A nested PCR method was used to amplify ABL kinase domain with first step primers BCR forward primer (TGACCAACTCGTGTGTGAAACTC) and ABL kinase reverse primer (TCCACTTCGTCTGAGATACTGGATT). Then a second-stage PCR on the produced PCR product was performed using ABL kinase forward primer (CGCAACAAGCCCACTGTCT) and reverse primer (TCCACTTCGTCTGAGATACTGGATT) with annealing temperature of 65°C (12). After performing purification steps using purification kit (QIA-GEN, Germany), the whole ABL kinase domain was sequenced in both directions using the sequencing kit (ABI BigDye terminator v3.1, USA) by the ABI 3130XL genetic analyzer. This region included 863bp (Gene Bank accession number M14752) and compared with the wild type ABL using Gene Runner last version for alignment.
3.4. Bioinformatics Analysis
The positions of reported ABL kinase domain variations were marked on a prepared plot for simple comparison between normal and mutated ABL to determine the mutations. An online program, Polyphen was used to predict the consequence of each mutation on the BCR-ABL protein structure.
4. Results
The demographic profile, disease characteristics and molecular analysis of the included patients are shown in Tables 2 and 3.
| Demographic | No. Patients, % |
|---|---|
| Male | 25 |
| Female | 14 |
| Average | 46 (22 - 70) |
| Male | 43 (22 - 64) |
| Female | 45 (30 - 70) |
| Demographic | Patients, % |
|---|---|
| 0 - 12 | 5 |
| 13 - 24 | 6 |
| 25 - 36 | 4 |
| 37 - 48 | 6 |
| 49 - 60 | 6 |
| > 61 | 11 |
| Chronic phase | 4 |
| Accelerated phase | 6 |
| Blast phase | 0 |
| Complete | 14 |
| Loss of HR | 28 |
| Absent | 6 |
| Present | 4 |
| G250E | 1 |
| L384M | 1 |
| Y253H | 1 |
| V379I | 1 |
a At the time of referral to the laboratory.
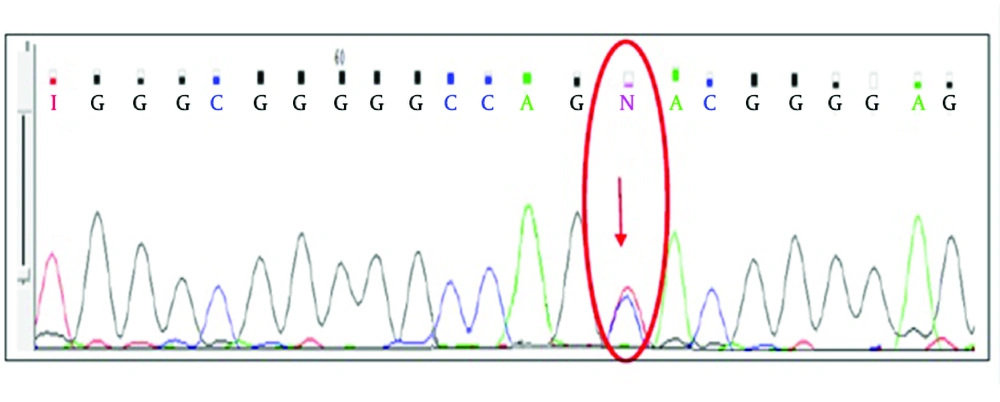
More than 80% of CML patients, referred to Payvand Clinical and Specialty Laboratory to follow-up, were in the first line TKI therapy. Among them, 39 of 135 (28.8%) who showed Imatinib resistance criteria, entered the study. Of 39 selected resistant patients, 25 (64.1%) were male and 14 (35.9%) female. The predominance of the disease was observed among males. Hematology tests for measuring main indexes like white blood cells (WBC), platelet and basophil were performed on taken peripheral blood samples to determine the hematologic response to the therapy. Real-time PCR was performed to estimate molecular response by specifying the BCR-ABL/ABL copy numbers on the synthesized c-DNA. Twenty-five (63.2%) patients were categorized as failure molecular response, four (10.2%) were in warning category and 10 (26.3%) in optimal response according to the 2013 ELN guideline (10). CHR was not seen in any of resistant patients classified in failure and warning groups of molecular response. The results of RFLP method performed on ABL amplified kinase domain for unveiling the proportion of mutated and unmutated alleles of four selected common mutations. According to the direct RFLP results, among 39 selected patients tested for the common named mutations, only one of them showed the Y253H mutation. Four (10.25%) of 39 resistant patients showed mutations in ABL kinase domain as sequenced the ABL domain. Among these, the Y253H mutation was shown in one patient by RFLP method and confirmed by direct sequencing (Figures 2 and 3).
Mutation at amino acid 253 is predicted to impair binding of Imatinib to BCR-ABL by disrupting van der Waals interaction (12). Nine CML resistant patients were randomly selected among 39 resistant patients to be sequenced the kinase domain of ABL gene. G250E and L384M mutations were found in ATP binding site (P-loop). Mutations within ATP binding site prevent Imatinib from binding to amino acid in ABL kinase domain by interrupting critical contact between Imatinib and the protein or inducing a conformation to which Imatinib cannot bind. Consequently, ATP binding site is not blocked and phosphate transfers and downstream transduction is performed properly (13). In addition, the results of one patient showed the V379I mutation between catalytic domain and activation site A-loop.
The mutations explained above, have a critical role in abrogating binding to BCR-ABL, reportedly. Although they are not certainly involved in disruption of Imatinib binding, their actual function is not identified (13)
5. Discussion
All patients with a mutation in ABL kinase domain showed treatment failure based on 2013 ELN recommendations for management of CML. BCR-ABL kinase domain mutations represented the major cause of resistance to TKI therapy (5). More than 90 various mutations occurred in patients with Imatinib resistance. Of these, clinical investigations demonstrated 15 amino acid substitutions accounting for 80 - 90% of cases representing Imatinib resistance mutations. In addition, seven mutated codons (G250E, Y253H, E255K, T315I, M351T, F359 and H396) are accounted for about 60 - 70% of the cases (5, 9). In this study, we considered a number of factors including lack of MMR, increase of BCR-ABL transcript level up to five fold, relapse in CHR or no CHR during therapy, being more than four years under TKI therapy, switching to 2nd or 3rd generations of TKI to ensure the high probability of having mutations in resistant patients.
Four different ABL kinase domain mutations, reported earlier were identified in four (10.25%) CML Imatinib resistant patients. Among the mutated patients, the V379I mutation occurred in a-56-year-old male who had been under therapy for 17 years. This patient exhibited CHR in blood specimen, while he did not achieve the MMR. The mutation L384M was detected in a patient with hematologic remission confirmed by platelet count up to 1100/L. MMR was not seen in this patient either. Increasing the BCR-ABL ratio up to 63% after five years of therapy showed that a mutation, which occurred at position 253, changed the amino acid Y to histidine. Furthermore, hematologic indexes showed no CHR in this case. The mutation G250E was seen at the beginning of the ABL sequence in a patient who was in accelerated phase (AP), at the time of study. In addition, both CHR and MMR were not achieved in the mentioned patient. Overall, 3 of 4 mutated patients were in accelerated phase at the time of study and just one of them was in chronic phase (CP). Consistent with the data, reported earlier from Imatinib resistant patients, detection of ABL kinase domain mutations was more frequent in patients in accelerated phase. Among patients with dose escalation up to 500 mg and 600 mg daily, no mutation was detected. Possibly, these mutations were previously present in these cases. However, their effect may be compensated by the dose escalation. Moreover, all patients with mutation showed treatment failure according to Real-time PCR results. Genomic mutations occurred more frequently in patients with high prognostic risk. Since these mutated patients were less likely to achieve optimal response to TKI, a hypothesis could be that the presence of a mutation is related to susceptibility for occurrence resistance due to genomic instability (5).
Based on reported causes of developing resistance, ABL KD mutations did not account for all cases with loss of response to therapy (5). This investigation suggests that other mechanisms causing resistance may be effective in some resistant patients. There may be some factors which were not considered in this analysis, but have an effective role on the development of mutations and response to therapy.
One of the study limitations was limited number of samples. Research on larger number of patients is suggested to obtain more reliable results. Due to the lack of information on ABL kinase domain mutations in Iranian population, it is difficult to conclude based on the study results, since the comparison of results with those of the other studies is not possible. Furthermore, our research was affected by the short period of the study and impossibility of follow-up in all cases. To draw reliable conclusions, more factors should be considered including patient adherence, more long term and effective follow-up and molecular and cytogenetic response comparison.
According to our testing results, mutations detected in ABL kinase domain were the cause of rising BCR-ABL/ABL ratio and occurring resistant in CML patients under first line tyrosine kinase therapy. Increasing BCR-ABL/ABL ratio with respect to international scale ELN in patients under Imatinib therapy is a mean of detecting Imatinib resistant patients. It seems that the pathway is round trip. In Y253H mutated patients, the BCR-ABL/ABL ratio was around 63% after five years therapy. In addition G250E was detected in a patient with three years therapy without decreasing the ratio under 40%. Moreover, hematologic and molecular responses, not seen in patient who hold L384M mutation and consumed the top limitation of Imatinib usage (8 tab per day) cannot overcome the increased BCR-ABL/ABL ratio under 0.1%. Therefore, regular assessment of molecular response may help physicians to discover the chance of remission or any new decision for changing therapeutic strategies.