1. Context
Breast cancer (BC) counts the most common cause of cancer related death among women. The studies reported about 235303 new cases of BC and about 40430 death records per year in the United States (1). BC is the result of collaboration of many variables including environmental, reproductive, lifestyle, and genetic related factors (2) and approximately 10% of BC cases are attributed to genetic factors (3).
Mutations in high-penetrance genes such as BRCA1, BRCA2, TP53 and PTEN are demonstrated to contribute in BC susceptibility (4). However, they are counted only for 25% of inherited BC risk (5). A combined polymorphism effect of moderate and common low risk genes can clear remaining hereditary BC risk (6). Base-excision repair (BER) genes are an important subcategory of DNA repair genes, which fix DNA base damage due to X-rays, oxygen radicals, and alkylating agents (7). BER pathway excise oxidized DNA bases when oxidized DNA damage occur (8). Mutations in BER genes by cumulative effect of endogenous and exogenous mutagens result in apoptosis or cell overgrowth and cancer. Known polymorphisms of DNA repair genes can be associated with the risk of many types of cancer such as lung (9), colorectal (10), bladder (11), and BC (12). On the other hand, there are evidences demonstrating no association or protective effect of BER genes with breast cancer (11, 13, 14).
X-ray repair cross-complementing group 1 gene (XRCC1) and 8-oxoguanine DNA glycosylase (OGG1) play an important role in base-excision repair pathway. XRCC1 acts in BER encoding scaffolding protein that assemble together proteins of the DNA repair complex (15, 16). Arg194Trp, Arg280His and Arg399Gln amino acid substitution are the most common studied single nucleotide polymorphisms of XRCC1. OGG1 encodes a DNA glycosylase, an important enzyme in the repair of 8-oxoguanine (17). Amino acid replacement of serine (Ser) with cysteine (Cys) at codon 326 (Ser326Cys, rs1052133) as a result of common C/G single nucleotide polymorphism (SNP) in exon 7 of hOGG1 gene is associated with decreased hOGG1 repair activity (18).
There are studies recommending variation in association of genetic polymorphisms and diseases in different ethnicities and geographical populations (19, 20). There is no consensus among researchers regarding association between XRCC1 (21-24) and OGG1 (12, 25, 26) with BC. A case-control study in the United States and Poland populations which evaluated association of BER genes with BC (27) showed no association for XRCC1 and OGG1 in both populations. While other studies (28) (29) suggested association of XRCC1 (codon 194, codon 280, and codon 399) and OGG1 (codon 326) with breast cancer.
2. Objectives
As such, we aimed to conduct a comprehensive systematic review to address the association of XRCC1 and OGG1 polymorphisms with breast cancer.
3. Data Sources
The systematic review was conducted according to the meta-analysis of observational studies in epidemiology (MOOSE) group’s criteria (30). To extract all studies around genetic polymorphisms and breast cancer, we performed a sensitive search in PubMed, PubMed central and ISI web of knowledge from their commencements until September 2014. The search was carried out using keywords “Breast Neoplasms”, “Breast Tumor”, “Human Mammary Neoplasm”, “Human Mammary Carcinoma”, “Cancer of Breast, Breast Cancer”, “Mammary Cancer”, “Malignant Neoplasm of Breast”, “Malignant Tumor of Breast”, “Breast Carcinoma”, “Cancer of the Breast”, “Ductal Carcinoma of Breast”, “Lobular Carcinoma of Breast”, “Medullary Breast Cancer”, “Breast Tumor” and “In Situ Breast Cancer” with Boolean “OR” between them combining “Polymorphism” with Boolean “AND” (each dataset detailed search string and retrieved results reported in Appendix 1). Search results were limited to human studies. We also performed contemporary hand search of reference lists of final retrieved studies, meta-analysis and systematic reviews.
4. Study Selection
All longitudinal cohorts (conventional cohort and historical cohort studies) and case-control studies (conventional case-control, case-cohort, nested case-control, matched case-control and unmatched case-control studies) investigating association of breast cancer with XRCC1 and OGG1 genes and their polymorphisms were eligible for initial inclusion. Also, all types (carcinomas, sarcomas) and grades (low, intermediate/moderate and high grade) of breast cancer were considered in this review. The criteria for exclusion of articles on the basis of title and abstract screening were: 1) Studies with no control group, 2) non-research articles (all type of letters, comments, and editorial), 3) animal studies, 4) case reports and case series studies, 5) considered occurrence of secondary/metastatic breast cancer or all-cause mortality as an outcome , 6) considered patients undergoing any type of intervention and 7) studies without breast cancer (pre/post-menopausal) as outcome. If the same population had been studied in two or more different studies (articles), we chose the most comprehensive and the most recent one.
5. Data Extraction
Two authors screened titles and abstracts and extracted all needed information from eligible studies after exclusion of duplicate titles. If there was a disagreement in each stage, three authors discussed conflicting results. EndNote X7 software was used to manage review and organize screening.
The following data were extracted from each study: name of first authors, date of publication, study design, source of controls, considered confounders in each models, genotyping methods, population ethnicity, total number of cases and controls, menopausal status of cases and controls, number of cases and controls according to genotypes and menopausal status, mean age of cases and controls, frequency of each genotypes separately and odds ratios for homozygote, heterozygote and combined model.
Because of the potential hazards of assessing the quality of studies using mechanical checklist and scoring the pre-considered item which is not made specifically for genetic epidemiology researches (31-34). For quality assessment procedure, we used four research methodological components causing bias for the association between gene polymorphisms and breast cancer risk, including source of controls sampling, population ethnicity, sample size of studies and menopausal status of cases and controls (Appendix 2 to 5). In order to minimize potential errors, considered data extracted on a former piloted Microsoft Excel worksheet, accompanied by predefined instructions for reviewers. In final stage, senior researcher re-checked extracted data. If clarifications and more information (or unavailable full texts) were needed, we contacted the first and corresponding author for additional data.
6. Results
6.1. Eligible Studies
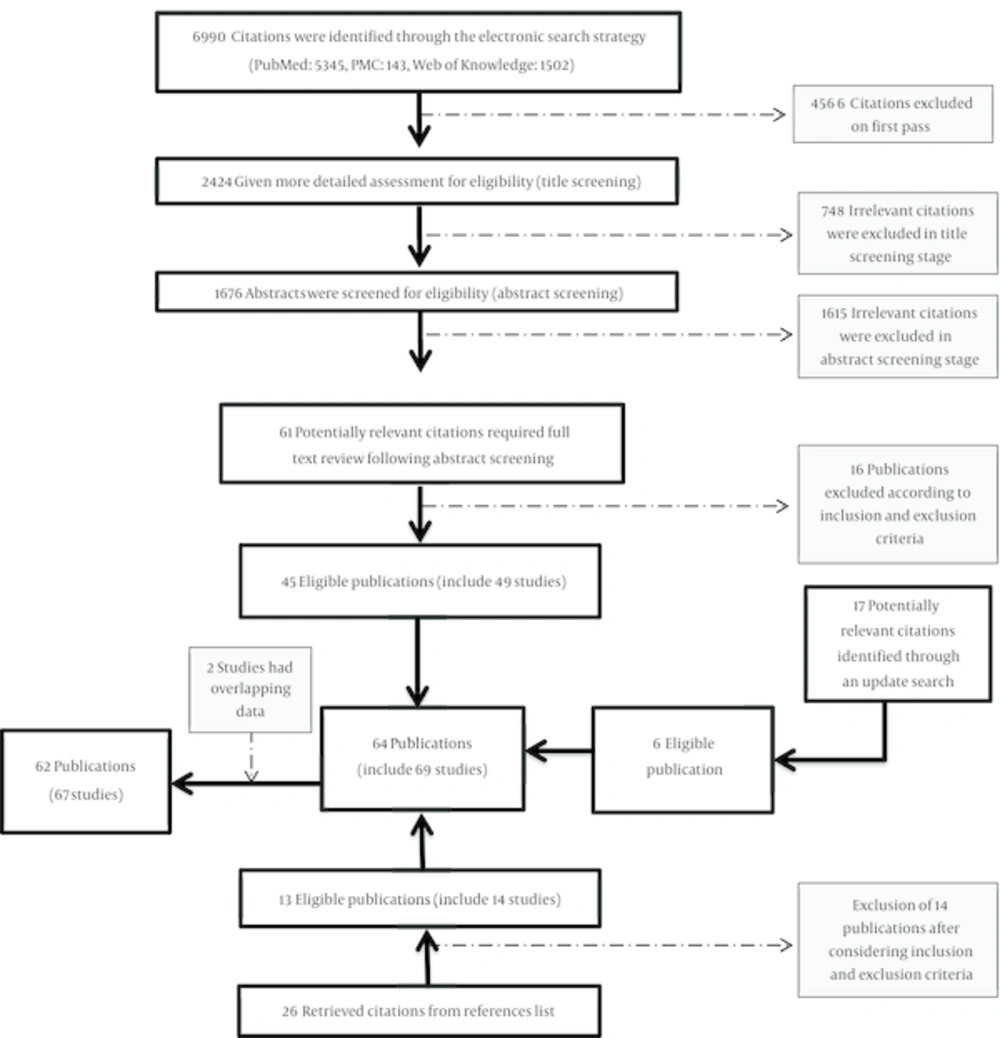
A total of 2424 titles were identified in initial electronic search. After final screening, 45 eligible publications (including 49 studies) through primary search, 13 publications (14 studies) through hand search of references list of retrieved citations and 6 through an update search of databases were retrieved (Figure 1). Two studies (35, 36) were excluded because of having overlapping population with two others (37, 38). Overall, 62 publications (67 studies) were included in this study according to inclusion and exclusion criteria (12, 21-23, 25-29)(37-89). General characteristics of all eligible studies separated for each gene are provided in Appendix 6 to 9. Different ethnicities populations in the same study were considered as separated study populations (five publications comprised of two populations with different ethnicities) (27, 43, 47, 68, 83). From these, 35 studies worked on Arg194Trp, 12 studies evaluated Arg280His, 56 assessed Arg399Gln, and 19 studies focused on Ser326Cys. All studies consist of retrospective confirmed breast cancer cases and cancer-free controls and were published during 2001 to 2015. Among retrieved studies, 17 also presented information on menopausal status and six studies were conducted on postmenopausal women. Considering geographical location, 24 studies were carried out in North America, 21 in Europe, 19 in Asia, 2 in Latin America and 2 in Africa. To have a better view of measure of association, ORs were recalculated for five studies which did not take wild type as reference category (46, 52, 56, 74, 87), four studies because of stratification were based on family history of breast cancer (22, 44, 48, 86) and one because of stratification was based on recreational activity (63).
6.2. Arg194Trp
General characteristics of eligible studies are exhibited in Appendix 6. The 35 studies on Arg194Trp consisted of 14,793 breast cancer cases and 15,409 controls. Sample size of the studies ranged from 74 to 2,833 individuals. The largest sample size belonged to Zhang’s study (27) and Sterpone’s study had the lowest sample size (38). Tryptophan (Trp) allele frequency ranged from 0.03 to 0.53 in cases and 0.02 to 0.57 in controls. Most of the studies (16 of 35) were conducted in the United States of America. Clinic (or hospital) based case-control studies were the most common design (19 of 35). In addition, polymerase chain reaction-restriction fragment length polymorphism method (PCR- RFLP) was the most used genotyping method (20 of 35).
Among these 35 researches, four studies (28, 39, 70, 71)28, 39, 70, 71) showed significant association between Arg194Trp polymorphism and breast cancer comparing Trp carriers versus wild type and two (21, 59) showed marginally significant association. On the contrary, one study displayed protective effects (66). ORs ranged from not calculable (NC) (not enough number of exposed individuals) to 8.74 (adjusted) and NC to 3.87 (unadjusted) for homozygote versus wild type, from 0.44 to 4.42 (adjusted) and 0.35 to 4.59 (unadjusted) for heterozygote versus wild type and from 0.62 to 4.28 (adjusted) and 0.35 to 4.571 (unadjusted) for Trp carriers versus wild type. After considering stratification for menopausal status, only one study (28) reported significant association for post menopause and none showed association for premenopausal women. Oppositely, Patel (69) showed protective effect of Arg194Trp polymorphism on postmenopausal women (Table 1).
| References | Menopausal Status | Trp/Trp Versus Arg/Arg | Arg/Trp Versus Arg/Arg | Trp/Trp and Arg/Trp Versus Arg/Arg | |||
|---|---|---|---|---|---|---|---|
| Adjusted OR (95% CI) | Unadjusted OR (95% CI) | Adjusted OR (95% CI) | Unadjusted OR (95% CI) | Adjusted OR (95% CI) | Unadjusted OR (95% CI) | ||
| Mix | - | NC | - | 0.64 (0.28 - 1.42) | 0.70 (0.30 - 1.50) | - | |
| Mix | - | NC | - | 0.75 (0.40 - 1.39) | 0.70 (0.40 - 1.30) | - | |
| Mix | 0.90 (0.48 - 1.67) | - | 1.10 (0.76 - 1.73) | - | - | 1.08 (0.71 - 1.62) | |
| Pre | 1.30 (0.57 - 2.95) | - | 1.30 (0.74 - 2.18) | - | - | 1.27 (0.74 - 2.18) | |
| Post | 0.50 (0.17 - 1.32) | - | 1.00 (0.50 - 1.86) | - | - | 0.83 (0.42 - 1.62) | |
| Mix | - | 1.33 (0.17 - 10.00) | - | 0.79 (0.61 - 1.03) | 0.79 (0.60 - 1.04) | 0.81 (0.62 - 1.05) | |
| Mix | 1.61 (0.10 - 26.10) | - | 0.95 (0.56 - 1.61) | - | - | 1.02 (0.61 - 1.71) | |
| Mix | NC | - | 1.47 (0.80 - 2.70) | - | 1.60 (0.89 - 2.87) | - | |
| Mix | - | NC | - | 1.10 (0.58 - 2.02) | - | 1.12 (0.60 - 2.03) | |
| Mix | - | NC | - | 0.41 (0.16 - 1.06) | - | 0.47 (0.19 - 1.17) | |
| Mix | - | NC | - | 1.27 (0.58 - 2.78) | - | - | |
| Mix | 2.78 (0.82 - 9.40) | 2.73 (0.81 - 9.20) | 1.89 (0.99 - 3.62) | 1.85 (1.01 - 3.38) | 2.04 (1.12 - 3.72) | 1.98 (1.13 - 3.48) | |
| Pre | - | - | - | - | 1.35 (0.55 - 3.25) | - | |
| Post | - | - | - | - | 2.75 (1.28 - 5.90) | - | |
| Post | - | - | - | - | 0.62 (0.40 - 0.95) | 0.66 (0.45 - 0.97) | |
| Mix | - | - | - | - | 0.93 (0.72 - 1.21) | - | |
| Pre | - | - | - | - | 0.78 (0.50 - 1.20) | - | |
| Post | - | - | - | - | 1.00 (0.72 - 1.38) | - | |
| Mix | - | NC | - | 1.69 (0.99 - 2.87) | 1.20 (0.72 - 2.00) | 1.12 (0.69 - 1.83) | |
| Mix | 0.70 (0.10 - 4.40) | - | 1.00 (0.70 - 1.30) | - | 1.00 (0.70 - 1.30) | - | |
| Mix | 0.90 (0.30 - 2.80) | - | 0.90 (0.70 - 1.20) | - | 0.90 (0.70 - 1.20) | - | |
| Mix | 0.98 (0.21 - 4.63) | 1.02 (0.23 - 4.57) | 1.23 (0.21 - 4.63) | 1.20 (0.77 - 1.87) | 1.21 (0.78 - 1.88) | 1.18 (0.77 - 1.81) | |
| Mix | 0.50 (0.20 - 1.40) | - | 0.90 (0.80 - 1.20) | - | - | 0.92 (0.73 - 1.15) | |
| Pre | 0.60 (0.00 - 10.30) | - | 1.20 (0.80 - 1.80) | - | - | 1.19 (0.81 - 1.77) | |
| Post | 0.50 (0.20 - 1.40) | - | 0.80 (0.60 - 1.00) | - | - | 0.75 (0.55 - 1.02) | |
| Mix | 1.65 (0.10 - 27.43) | 2.00 (0.12 - 32.14) | 1.48 (0.95 - 2.32) | 1.46 (0.94 - 2.26) | 1.49 (0.95 - 2.31) | 1.47 (0.95 - 2.27) | |
| Pre | NC | 0.05 (0.00 - 4.04) | 0.78 (0.16 - 3.68) | 0.83 (0.18 - 3.69) | 0.78 (0.16 - 3.68) | 0.81 (0.18 - 3.63) | |
| Post | - | - | - | - | - | - | |
| Mix | - | 0.95 (0.36 - 2.46) | - | 1.02 (0.82 - 1.28) | - | 1.02 (0.81 - 1.28) | |
| Mix | - | 0.41 (0.08 - 2.08) | - | 0.35 (0.18 - 0.65) | - | 0.35 (0.19 - 0.64) | |
| Mix | 0.70 (0.42 - 1.14) | 0.75 (0.47 - 1.20) | 1.08 (0.81 - 1.44) | 1.08 (0.82 - 1.41) | 1.01 ( 0.77 - 1.32) | ||
| Pre | 0.88 (0.45 - 1.73) | - | 1.23 (0.83 - 1.82) | - | - | 1.15 (0.80 - 1.65) | |
| Post | 0.55 (0.26 - 1.16) | - | 0.92 (0.60 - 1.42) | - | - | 0.85 (0.56 - 1.28) | |
| Mix | 8.74 (0.97 - 78.23) | - | 1.21 (0.74 - 1.97) | - | - | 1.34 (0.82 - 2.18) | |
| Mix | NC | - | 0.44 (0.12 - 1.67) | - | - | 0.82 (0.23 - 2.73) | |
| Post | - | 0.71 (0.33 - 1.54) | - | 0.74 (0.36 - 1.50) | - | 0.73 (0.37 - 1.41) | |
| Mix | 1.37 (0.87 - 2.17) | 1.36 (0.88 - 2.14) | 1.12 (0.84 - 1.47) | 1.09 (0.83 - 1.44) | 1.20 (0.87 - 1.62) | 1.17 (0.89 - 1.53) | |
| Mix | - | NC | - | 1.82 (0.43 - 7.66) | - | - | |
| Mix | - | - | - | - | 1.17 (0.61 - 2.25) | - | |
| Mix | 1.16 (0.84 - 1.59) | - | 1.19 (0.99 - 1.44) | - | 1.19 (0.99 - 1.41) | - | |
| Mix | - | NC | - | 4.57 (1.47 - 14.21) | - | 4.57 (1.47 - 14.21) | |
| Mix | - | NC | - | 1.91 (1.15 - 3.14) | - | 1.94 (1.15 - 3.32) | |
| Mix | - | 0.99 (0.66 - 1.48) | - | 0.93 (0.73 - 1.18) | 0.94 (0.75 - 1.18) | 0.94 (0.74 - 1.18) | |
| Post | - | - | - | - | - | 0.91 (0.62 - 1.32) | |
| Mix | 3.76 (1.07 - 13.17) | 3.87 (1.24 - 12.05) | 4.42 (2.08 - 9.41) | 4.59 (2.33 - 9.06) | 4.28 (2.11 - 8.70) | 4.45 (2.35 - 8.45) | |
| Mix | - | 0.89 (0.34 - 2.28) | - | 0.59 (0.26 - 1.35) | - | 0.68 (0.29 - 1.57) | |
| Mix | - | 1.41 (0.64 - 3.14) | - | 1.01 (0.71 - 1.44) | - | 1.06 (0.76 - 1.48) | |
Abbreviations: CI, confidence interval; NC, not calculable; OR, odds ratio.
aReferences category for calculation of odds ratio.
bORs calculated by authors via Stata software (version 13).
6.3. Arg280His
A total of 7,716 cases and 7,370 controls were studied for Arg280His. Zhang’s study (27) had the largest sample size (n = 2,802) and Smith’s study (83) (this study consists of two different ethnic populations and here the African American population was studied) had the lowest (n = 128). Histidine (His) allele had the least frequency among studies population comparing to other three alleles. His allele frequency ranged from 0.03 to 0.11 in cases and from 0.03 to 0.14 in controls. Six of studies (4 publications) were conducted in the United States of America (27, 68, 83, 89). Seven studies (six publications) had a clinic-based design and five (four publications) had a population-based design. Genotyping of four studies (28, 59, 60, 64) was carried out by PCR-RFLP (Appendix 7).
Results of Arg280His showed significant association between Arg280His polymorphism and breast cancer comparing His carriers versus wild type only in one study (67) with OR = 1.82 (1.06 - 3.15). ORs ranged from NC to 1.69 (adjusted) and NC to 4.68 (unadjusted) for homozygote versus wild type, from 0.55 to 1.80 (adjusted) and 0.56 to 1.13 (unadjusted) for heterozygote versus wild type and from 0.61 to 1.3 (adjusted) and 0.63 to 1.82 (unadjusted) for His carriers versus wild type. Stratification for menopausal status revealed marginally significant association in one study (29) (OR = 1.71 (0.94 - 3.10); P = 0.07) and protective association of Arg280His polymorphism in another study (28) (OR = 0.26 (0.1 - 0.66)) for post-menopausal women (Table 2).
| References | Menopausal Status | His/His Versus Arg/Arg | Arg/His Versus Arg/Arg | His/His and Arg/His Versus Arg/Arg | |||
|---|---|---|---|---|---|---|---|
| Adjusted OR (95% CI) | Unadjusted OR (95% CI) | Adjusted OR (95% CI) | Unadjusted OR (95% CI) | Adjusted OR (95% CI) | Unadjusted OR (95% CI) | ||
| Mix | NC | - | 1.80 (1.04 - 3.08) | - | - | 1.82 (1.06 - 3.15) | |
| Mix | 1.69 (0.29 - 9.63) | 1.79 (0.32 - 10.05) | 0.55 (0.29 - 1.03) | 0.56 (0.29 - 1.05) | 0.61 (0.33 - 1.12) | 0.63 (0.35 - 1.15) | |
| Pre | - | - | - | - | 1.44 (0.62 - 3.70) | - | |
| Post | - | - | - | - | 0.26 (0.10 - 0.66) | - | |
| Mix | - | NC | - | 1.13 (0.78 - 1.63) | 1.15 (0.80 - 1.66) | - | |
| Mix | 1.10 (0.10 - 18.00) | - | 1.30 (0.80 - 2.00) | - | 1.30 (0.80 - 1.90) | - | |
| Mix | NC | - | 1.20 (0.90 - 1.60) | - | 1.20 (0.90 - 1.60) | - | |
| Mix | 1.00 (0.20 - 4.30) | 1.10 (0.80 - 1.40) | 1.10 (0.85 - 1.44) | ||||
| Pre | 0.30 (0.00 - 3.50) | - | 1.00 (0.60 - 1.40) | - | - | 0.94 (0.62 - 1.43) | |
| Post | 2.20 (0.20 - 21.20) | - | 1.10 (0.80 - 1.60) | - | - | 1.14 (0.80 - 1.63) | |
| Mix | - | 4.68 (1.01 - 21.70) | - | 0.89 (0.71 - 1.11) | - | 0.92 (0.73 - 1.15) | |
| Mix | - | - | - | - | 1.30 (0.88 - 1.93) | 1.09 (0.76 - 1.58) | |
| Pre | - | - | - | - | 0.99 (0.58 - 1.70) | - | |
| Post | - | - | - | - | 1.71 (0.94 - 3.10) | - | |
| Mix | NC | - | 0.72 (0.43 - 1.21) | - | - | 0.70 (0.40 - 1.19) | |
| Mix | NC | - | 0.66 (0.14 - 3.05) | - | - | - | |
| Mix | - | - | - | - | 0.87 (0.38 - 2.00) | - | |
| Mix | 0.50 (0.21 - 1.18) | - | 1.02 (0.81 - 1.28) | - | 0.98 (0.78 - 1.22) | - | |
Abbreviations: CI, confidence interval; NC, not calculable; OR, odds ratio.
aReferences category for calculation of odds ratio.
bORs calculated by authors via Stata software.
6.4. Arg399Gln
Arg399Gln was the most popularly evaluated polymorphism and 27167 cases and 31998 controls were studied to estimate association between Arg399Gln polymorphism and breast cancer. The sample sizes ranged from 74 to 7,25 individuals in which the largest and smallest studies were carried out by Sapkota (79) and Sterpone (38), respectively. Glutamine (Gln) allele frequency ranged from 0.14 to 0.55 in cases and 0.11 to 0.57 in controls. 19 of 56 studies (16 publications) were conducted in the United States of America. Thirty-one studies used PCR-RFLP method for genotyping and thirty-three studies had clinic based study design (Appendix 8).
Among studies analyzing Arg399Gln, seven (28, 38, 47, 61, 71, 80, 86) showed significant association between Arg399Gln polymorphism and breast cancer comparing Gln carriers versus wild type. Although six studies (27, 29, 53, 62, 64, 66) showed a borderline association, just one (64) had P < 0.05. Conversly, Zipprich et al. (89) revealed protective effect of Arg399Gln on breast cancer risk (OR = 0.64 (0.41 - 1.00)). Calculated ORs ranged from 0.44 to 4.42 (adjusted) and 0.25 to 4.40 (unadjusted) for homozygote versus wild type, from 0.65 to 2.64 (adjusted) and 0.65 to 4.8 (unadjusted) for heterozygote versus wild type and from 0.64 to 2.89 (adjusted) and 0.60 to 4.66 (unadjusted) for Gln carriers versus wild type. Arg399Gln polymorphism showed to have an association with breast cancer in one study for Premenopausal women (28) and three for postmenopausal woman (61, 72, 74). Nonetheless, one study showed protective effect of Arg399Gln polymorphism for breast cancer risk (57). Details of extracted ORs are reported in Table 3.
| References | Menopausal Status | Gln/Gln Versus Arg/Arg | Arg/Gln Versus Arg/Arg | Gln/Gln and Arg/Gln Versus Arg/Arg | |||
|---|---|---|---|---|---|---|---|
| Adjusted OR (95% CI) | Unadjusted OR (95% CI) | Adjusted OR (95% CI) | Unadjusted OR (95% CI) | Adjusted OR (95% CI) | Unadjusted OR (95% CI) | ||
| Mix | - | 2.11 (0.52 - 9.99) | - | 1.54 (1.03 - 2.32) | 1.70 (1.10 - 2.40) | - | |
| Mix | - | 0.84 (0.52 - 1.33) | - | 1.12 (0.81 - 1.54) | 1.00 (0.80 - 1.40) | - | |
| Mix | 2.40 (1.20 - 4.72) | 0.80 (0.51–1.16) | 0.96 (0.63 - 1.44) | ||||
| Pre | 3.80 (1.44 - 10.30) | 1.20 (0.72–2.10) | 1.51 (0.87 - 2.60) | ||||
| Post | 1.40 (0.50 - 3.91) | 0.40 (0.19–0.74) | 0.50 (0.25 - 0.98) | ||||
| Mix | 1.03 (0.77 - 1.37) | 1.06 (0.81 - 1.39) | 1.07 (0.88–1.30) | 1.06 (0.88 - 1.27) | - | 1.04 (0.88 - 1.24) | |
| Mix | 1.00 (0.57 - 1.76) | - | 0.96 (0.66–1.40) | - | - | 0.91 (0.64 - 1.29) | |
| Mix | 1.20 (0.85 - 1.69) | 1.22 (0.87 - 1.71) | 0.90 (0.76–1.08) | 0.92 (0.77 - 1.09) | 1.00 (0.84 - 1.18) | ||
| Pre | 1.19 (0.78 - 1.80) | 1.20 (0.79 - 1.81) | 0.79 (0.63–0.98) | 0.79 (0.63 - 0.98) | 0.90 (0.73 - 1.10) | ||
| post | 1.13 (0.61 - 2.11) | 1.19 (0.65 - 2.17) | 1.19 (0.87–1.62) | 1.21 (0.90 - 1.64) | 1.20 (0.90 - 1.62) | ||
| Mix | 1.07 (0.58 - 1.96) | - | 1.02 (0.70–1.51) | - | 1.03 (0.71 - 1.49) | - | |
| Mix | - | 1.09 (0.54 - 2.16) | - | 0.81 (0.53 - 1.25) | - | 0.86 (0.57 - 1.29) | |
| Mix | - | 1.26 (0.61 - 2.61) | - | 0.88 (0.53 - 1.47) | - | 0.96 (0.59 - 1.56) | |
| Mix | 0.88 (0.57 - 1.37) | 0.92 (0.60 - 1.41) | 0.91 (0.67-1.23) | 0.92 (0.68 - 1.24) | - | 0.92 (0.68 - 1.23) | |
| Mix | - | 0.89 (0.46 - 1.72) | - | 1.10 (0.75 - 1.61) | - | 1.06 (0.73 - 1.52) | |
| Mix | 1.92 (0.79 - 4.69) | 0.99 (0.61 - 1.60) | 1.11 (0.71 - 1.75) | ||||
| Mix | 2.69 (1.10 - 6.57) | 2.66 (1.10 - 6.40) | 2.04 (1.16-3.58) | 2.02 (1.16 - 3.49) | 2.18 (1.30 - 3.66) | 2.14 (1.29 - 3.58) | |
| Pre | 6.32 (2.90 - 13.73) | ||||||
| Post | 0.46 (0.36 - 1.57) | ||||||
| Mix | - | 0.89 (0.26 - 2.85) | - | 0.84 (0.45 - 1.58) | - | 0.85 (0.47 - 1.54) | |
| Mix | 1.39 (0.84 - 2.29) | - | 1.24 (0.93–1.65) | - | 1.26 (0.96 - 1.66) | - | |
| Post | 1.27 (0.79 - 2.02) | 1.08 (0.71 - 1.63) | 1.01 (0.73–1.39) | 0.95 (0.71 - 1.27) | 1.06 (0.78 - 1.44) | 0.98 (0.75 - 1.28) | |
| Mix | 0.97 (0.73 - 1.29) | 1.08 (0.90-1.29) | 1.06 (0.89 - 1.26) | ||||
| Pre | 1.03 (0.62 - 1.70) | 1.28 (0.93-1.75) | 1.23 (0.91 - 1.66) | ||||
| Post | 1.01 (0.71 - 1.44) | 0.99 (0.79-1.24) | 0.99 (0.80 - 1.24) | ||||
| Mix | 1.22 (0.73 - 2.06) | 1.11 (0.67 - 1.83) | 1.64 (1.00–2.69) | 1.48 (0.92 - 2.38) | - | 1.24 (0.88 - 1.75) | |
| Mix | - | 1.10 (0.48 - 2.53) | - | 0.98 (0.58 - 1.65) | - | 1.00 (0.58 - 1.69) | |
| Mix | 1.80 (0.80 - 3.80) | - | 1.10 (0.90-1.50) | - | 1.20 (0.80 - 1.50) | - | |
| Mix | 1.00 (0.80 - 1.30) | - | 1.10 (0.90-1.30) | - | 1.10 (0.90 - 1.30) | - | |
| Mix | 0.91 (0.48 - 1.72) | 0.91 (0.49 - 1.70) | 1.29 (0.84–1.97) | 1.29 (0.85 - 1.95) | 1.20 (0.80 - 1.79) | 1.19 (0.80 - 1.77) | |
| Mix | 1.01 (0.60 - 1.70) | 0.79 (0.57-1.08) | 0.82 (0.61 - 1.11) | ||||
| Pre | 0.83 (0.55 - 1.28) | ||||||
| Post | 0.82 (0.54 - 1.25) | ||||||
| Mix | 0.90 (0.80 - 1.10) | 1.10 (0.90-1.20) | 1.03 (0.92 - 1.15) | ||||
| Pre | 0.80 (0.60 - 1.10) | 1.00 (0.80-1.20) | 0.92 (0.77 - 1.11) | ||||
| Post | 1.10 (0.80 - 1.30) | 1.10 (0.90-1.30) | 1.09 (0.95 - 1.26) | ||||
| Mix | 1.10 (0.90 - 1.40) | 1.10 (1.00-1.30) | 1.10 (0.97 - 1.25) | ||||
| Pre | 1.00 (0.70 - 1.50) | 1.20 (0.90-1.50) | 1.12 (0.88 - 1.44) | ||||
| Post | 1.10 (0.80 - 1.30) | 1.10 (0.90-1.30) | 1.08 (0.92 - 1.25) | ||||
| Mix | 0.95 (0.57 - 1.53) | 0.65 (0.47 - 0.90) | 1.64 (1.22 - 2.21) | ||||
| Mix | 0.87 (0.50 - 1.49) | 0.80 (0.47 - 1.36) | 0.87 (0.62–1.22) | 0.83 (0.60 - 1.16) | 0.87 (0.63 - 1.20) | 0.83 (0.60 - 1.13) | |
| Pre | 0.52 (0.14 - 2.61) | 0.65 (0.14 - 3.04) | 0.49 (0.17–1.42) | 0.49 (0.17 - 1.35) | 0.50 (0.18 - 1.32) | 0.52 (0.20 - 1.32) | |
| Post | |||||||
| Mix | - | 3.54 (0.69 - 10.65) | - | 1.52 (0.51 - 4.59) | - | 2.05 (0.76 - 5.61) | |
| Mix | - | 0.95 (0.39 - 2.27) | - | 1.25 (0.68 - 2.29) | - | 1.17 (0.67 - 2.04) | |
| Mix | - | 0.90 (0.68 - 1.18) | - | 0.95 (0.80 - 1.14 ) | - | 0.94 (0.79 - 1.11) | |
| Mix | - | 2.91 (1.66 - 5.10) | - | 0.91 (0.56 - 1.50) | - | 1.41 (0.90 - 2.19) | |
| Mix | 1.80 (0.90 - 3.61) | 2.01 (1.02 - 3.94) | 0.76 (0.47–1.20) | 0.76 (0.49 - 1.18) | 0.93 (0.61 - 1.44) | 0.95 (0.63 - 1.42) | |
| Pre | 1.93 (0.81 - 4.62) | 1.87 (0.81 - 4.37) | 0.88 (0.46–1.67) | 0.84 (0.45 - 1.56) | 1.09 (0.61 - 1.96) | 1.06 (0.60 - 1.85) | |
| Post | 1.20 (0.23 - 6.07) | 1.03 (0.21 - 5.05) | 0.48 (0.18–1.31) | 0.46 (0.17 - 1.21) | 0.57 (0.22 - 1.48) | 0.53 (0.21 - 1.34) | |
| Mix | 1.80 (0.99 - 3.29) | 1.74 (0.99 - 3.10) | 1.20 (0.90–1.60) | 1.17 (0.89 - 1.53) | 1.23 (0.94 - 1.61) | ||
| Pre | 1.45 (0.60 - 3.52) | 1.05 (0.71–1.55) | (0.75 - 1.57) | ||||
| Post | 1.84 (0.79 - 4.28) | 1.30 (0.84–2.00) | 1.40 (0.93 - 2.11) | ||||
| Mix | 0.93 (0.56 - 1.54) | - | 1.01 (0.74-1.40) | - | 1.00 (0.73 - 1.35) | 1.03 (0.75 - 1.40) | |
| Mix | 2.19 (0.09 - 52.25) | - | 1.13 (0.44-2.91) | - | 1.18 (0.47 - 2.96) | 1.33 (0.53 - 3.29) | |
| Post | - | 0.89 (0.42 - 1.84) | - | 1.09 (0.54 - 2.18) | - | 0.99 (0.52 - 1.88) | |
| Mix | - | 1.58 (0.99-2.52) | - | 1.60 (1.15 - 2.23) | - | 1.59 (1.17 - 2.16) | |
| Mix | 0.67 (0.36 - 1.25) | 0.67 (0.40 - 1.12) | 0.73 (0.48-1.10) | 0.85 (0.60 - 1.19) | 0.80 (0.57 - 1.13) | ||
| Mix | - | 0.94 (0.43 - 1.93) | - | 0.72 (0.44 - 1.20) | - | 0.77 (0.48 - 1.23) | |
| Mix | 1.31 (0.83 - 1.96) | 1.26 (0.82 - 1.93) | 1.05 (0.81–1.38) | 1.03 (0.78 - 1.36) | 1.12 (0.87 - 1.42) | 1.07 (0.82-1.40) | |
| Post | - | 2.79 (1.51 - 5.21) | - | 1.29 (0.83 - 1.99) | - | 1.59 (1.06 - 2.40) | |
| Mix | - | 0.25 (0.04 - 1.30) | - | 0.73 (0.30 - 1.40) | - | 0.60 (0.30 - 1.30) | |
| Mix | - | 4.40 (1.13 - 17.08) | - | 4.80 (1.56 - 14.78) | - | 4.66 (1.47 - 15.25) | |
| Mix | 0.44 (0.18 - 1.11) | - | 0.65 (0.39-1.10) | - | 0.64 (0.41 - 1.00) | - | |
| Mix | 0.91 (0.66 - 1.27) | - | 0.87 (0.72–1.05) | - | 0.88 (0.74 - 1.05) | - | |
| Mix | |||||||
| Pre | 0.79 (0.44 - 1.42) | 0.88 (0.62 – 1.24) | 0.86 (0.62 - 1.20) | ||||
| Post | 1.31 (0.94 - 1.83) | 1.22 (0.99 – 1.51) | 1.24 (1.01 - 1.51) | ||||
| Mix | 1.60 (0.60 - 4.10) | - | 1.70 (0.90 – 3.10) | - | 1.70 (0.90 - 2.90) | - | |
| Mix | - | 1.25 (0.46 - 3.39) | - | 0.94 (0.51 - 1.72) | - | 1.00 (0.57 - 1.75) | |
| Mix | - | 0.86 (0.46 - 1.60) | - | 1.16 (0.76-1.78) | - | 1.08 (0.70 - 1.65) | |
| Mix | 1.11 (0.94 - 1.31) | - | 1.12 (1.00 - 1.25) | - | - | - | |
| Mix | - | 2.69 (1.71 - 4.31) | - | 0.89 (0.70 - 1.14) | 1.10 (0.88 - 1.37) | 1.09 (0.87 - 1.38) | |
| Mix | 2.18 (1.06 - 4.50) | 1.52 (1.01 - 2.31) | 1.67 (1.12 - 2.47) | ||||
| Pre | 0.77 (0.20 - 2.91) | 1.53 (0.66-3.51) | 1.33 (0.60 - 2.91) | ||||
| Post | 2.57 (0.96 - 6.89) | 1.84 (1.08-3.15) | 1.94 (1.16 - 3.24) | ||||
| Post | - | - | - | - | - | 1.11 (0.87 - 1.42) | |
| Mix | 4.42 (1.28 - 15.31) | 3.56 (1.22 - 10.39) | 2.64 (1.20 – 5.08) | 2.62 (1.35 - 5.08) | 2.89 (1.35 - 6.21) | 2.77 (1.47 - 5.24) | |
| Mix | 1.76 (1.21 - 2.57) | - | 1.17 (0.92 – 1.51) | - | 1.29 (1.02 - 1.63) | - | |
| Mix | - | 2.71 (1.44 - 5.10) | - | 1.10 (0.80 - 1.51) | - | 1.27 (0.94 - 1.72) | |
Abbreviations: CI, confidence interval; OR, odds ratio; USA, United States of America.
aReferences category for calculation of odds ratio.
bORs calculated by authors via Stata software (version 13).
6.5. Ser326Cys
A total of 9,417 cases and 11,087 controls were founded to be considered for Ser326Cys. Respectively, Roberts’ study (72) and Sterpone’s (38) study had the largest and smallest sample sizes of 2,941 and 74 individuals. Cysteine (Cys) allele was the most prevalent allele in this study and Cys frequency ranged from 0.21 to 0.63 in cases and 0.21 and 0.68 in controls. About half of studies of Ser326Cys polymorphism were conducted in Asia, particularly on Chinese populations. Design of 13 studies (12 publications) was clinic based (Appendix 9).
Among studies focusing on Ser326Cys, none showed significant association between Ser326Cys polymorphism and breast cancer comparing Cys carriers to wild type. Calculated ORs ranged from 0.69 to 1.70 (adjusted) and 0.22 to 1.46 (unadjusted) for homozygote versus wild type, from 0.82 to 1.20 (adjusted) and 0.33 to 1.37 (unadjusted) for heterozygote versus wild type and from 0.76 to 1.21 (adjusted) and 0.28 to 1.28 (unadjusted) for Cys carriers versus wild type. Stratification for menopausal status exhibited no significant association between Ser326Cys and breast cancer in pre and post-menopausal women, but Romanowicz-Makowska (75) showed protective effect in post-menopausal women (OR = 0.49 (0.24 - 0.98)) (Table 4).
| First Author | Menopausal Status | Cys/Cys Versus Ser/Ser | Ser /Cys Versus Ser/Ser | Cys/Cys and Ser /Cys Versus Ser/Ser | |||
|---|---|---|---|---|---|---|---|
| Adjusted OR (95% CI) | Unadjusted OR (95% CI) | Adjusted OR (95% CI) | Unadjusted OR (95% CI) | Adjusted OR (95% CI) | Unadjusted OR (95% CI) | ||
| Mix | 1.20 (0.71 - 2.00) | - | 0.90 (0.59 - 1.53) | - | - | 0.94 (0.59 - 1.49) | |
| Pre | 1.10 (0.57 - 2.22) | - | 1.00 (0.56 - 1.93) | - | - | 1.03 (0.55 - 1.91) | |
| Post | 1.10 (0.46 - 2.45) | - | 0.70 (0.34 - 1.63) | - | - | 0.77 (0.37 - 1.64) | |
| Mix | 1.70 (0.93 - 2.96) | - | 1.20 (0.76 - 1.95) | - | - | 1.28 (0.81 - 2.02) | |
| Pre | 1.90 (0.75 - 4.90) | - | 1.30 (0.63 - 2.69) | - | - | 1.32 (0.65 - 2.66) | |
| Post | 1.30 (0.61 - 2.98) | - | 1.20 (0.64 - 2.42) | - | - | 1.23 (0.65 - 2.34) | |
| Post | - | 0.98 (0.52 - 1.86) | - | 0.84 (0.64 - 1.10) | - | 0.85 (0.64 - 1.13) | |
| Mix | - | 0.82 (0.42 - 1.61) | - | 0.90 (0.48 - 1.71) | - | 0.86 (0.48 - 1.58) | |
| Mix | 1.06 (0.83 - 1.35) | - | 1.17 (0.93 - 1.47) | - | - | 1.10 (0.88 - 1.38) | |
| Pre | 0.99 (0.74 - 1.34) | - | 1.08 (0.81 - 1.43) | - | - | 1.04 (0.79 - 1.37) | |
| Post | 1.18 (0.77 - 1.81) | - | 1.37 (0.91 - 2.07) | - | - | 1.26 (0.84 - 1.88) | |
| Mix | 1.01 (0.68 - 1.52) | 0.99 (0.66 - 1.47) | 1.06 (0.88 - 1.28) | 1.04 (0.87 - 1.24) | - | 1.02 (0.86 - 1.22) | |
| Mix | 1.00 (0.70 - 1.40) | - | 1.00 (0.80 - 1.20) | - | - | 0.98 (0.83 - 1.14) | |
| Pre | 0.90 (0.50 - 1.50) | - | 1.10 (0.80 - 1.40) | - | - | 1.02 (0.78 - 1.32) | |
| Post | 1.20 (0.70 - 1.90) | - | 1.00 (0.80 - 1.20) | - | - | 1.01 (0.82 - 1.25) | |
| Post | - | 0.62 (0.28 - 1.38) | - | 0.40 (0.18 - 0.87) | - | 0.49 (0.24 - 0.98) | |
| Mix | 1.42 (0.97 - 2.09) | 1.46 (1.02 - 2.10) | 0.95 (0.67 - 1.34) | 0.99 (0.72 - 1.37) | - | 1.14 (0.83 - 1.56) | |
| Pre | 1.13 (0.67 - 1.91) | - | 0.92 (0.58 - 1.48) | - | - | 1.12 (0.72 - 1.73) | |
| Post | 2.05 (1.14 - 3.69) | - | 0.97 (0.58 - 1.61) | - | - | 1.17 (0.72 - 1.89) | |
| Mix | - | 0.22 (0.04 - 1.04) | - | 0.33 (0.06 - 1.40) | - | 0.28 (0.05 - 1.10) | |
| Mix | 1.09 (0.74 - 1.59) | 1.09 (0.74 - 1.63) | 0.97 (0.66 - 1.42) | 0.99 (0.67 - 1.47) | 1.04 (0.80 - 1.46) | 1.02 (0.71 - 1.48) | |
| Mix | - | 0.83 (0.10 - 6.65) | - | 1.37 (0.53 - 3.56) | - | 1.03 (0.46 - 3.64) | |
| Mix | - | - | - | - | - | - | |
| Pre | 1.17 (0.53 - 2.54) | - | 1.00 (0.72 - 1.38) | - | 1.01 (0.74 - 1.38) | - | |
| Post | 1.19 (0.77 - 1.85) | - | 0.97 (0.79 - 1.19) | - | 0.99 (0.82 - 1.21) | - | |
| Mix | - | 0.83 (0.53 - 1.30) | - | 0.95 (0.66 - 1.38) | 0.90 (0.64 - 1.27) | 0.91 (0.64 - 1.30) | |
| Mix | - | 1.34 (0.97 - 1.84) | - | 1.10 (0.82 - 1.49) | - | 1.19 (0.88 - 1.60) | |
| Pre | - | 1.87 (1.14 - 3.06) | - | 1.08 (0.68 - 1.72) | - | 1.34 (0.84 - 2.15) | |
| Post | - | 1.18 (0.73 - 1.89) | - | 1.02 (0.66 - 1.57) | - | 1.07 (0.69 - 1.65) | |
| Mix | 0.69 (0.40 - 1.19) | - | 0.82 (0.48 - 1.38) | - | 0.76 (0.46 - 1.24) | - | |
| Pre | 0.41 (0.14 - 1.18) | - | 0.58 (0.21 - 1.64) | - | 0.49 (0.19 - 1.27) | - | |
| Post | 0.83 (0.40 - 1.70) | - | 1.01 (0.51 - 1.99) | - | 0.93 (0.49 - 1.76) | - | |
| Post | - | - | - | - | - | 0.98 (0.77 - 1.25) | |
| Mix | - | - | - | - | 1.82 (1.31 - 2.52) | - | |
| Mix | - | 0.93 (0.34 - 2.51) | - | 1.02 (0.45 - 2.34) | - | 1.00 (0.42 - 2.37) | |
Abbreviations: CI, confidence interval; OR, odds ratio.
aReferences category for calculation of odds ratio.
bORs calculated by authors via Stata software (version 13).
cCys/Cys versus Ser /Cys and Ser/Ser; Cys/Cys as reference category.
As part of a systematic review to address variations seen in the association between polymorphic genes involved in base excision repair pathway and risk of breast cancer, we identified all studies indexed in PubMed and Web of Sciences in which the risk of barest cancer for carriers of susceptibility alleles of XRCC1 and OGG1 genes were investigated. Collectively, there were 62 studies assessing the BER gene polymorphism in different population providing a wealth of information to look at the cause of variation in allele frequency as well as the magnitude of association between this polymorphic genes and breast cancer.
Etiology of breast cancer is characterized with major influence of endogenic estrogen exposure and gens involved in cancer susceptibility or genes involved in different pathways of estrogen metabolism (90). The lifestyle and environmental factors play important role in modifying the magnitude of estrogen exposure in a background of several susceptibility genes (91). The highly penetrate genes of BRCA1 and BRCA2 accounts for up to 25% of inherited breast cancer (where not more than 10% of breast cancer are considered as familial and inherited) (92, 93). The involvement of low-penetrate susceptibility polymorphic genes involved in the pathophysiologic pathways of breast tumor genesis play major role in sporadic breast cancer (94). The polymorphic genes can be involved in several cellular mechanisms especially estrogen metabolism pathways, detoxification of xenobiotic, cell adhesion, and DNA damage and repair signaling. While polymorphic genes involved in estrogen metabolism are very specific to breast cancer susceptibility, polymorphic genes involved in DNA repair and damage play an important role in the etiology of many cancers including breast (95). Among the many polymorphic genes involved in the etiology of breast cancer, Base excision repair polymorphic genes shows especially important role as a mechanism involved in repairing endogenous driven DNA damage, which may originate from wide range of normal metabolism. The magnitude of association (expressed as odds ratio in case-control and rate ration in cohort studies) between low penetrance genes and risk of cancer is very small and barely ranges more than 1.5. From population standpoint, the small magnitude of association seen between the susceptibility allele and risk of cancer is aggregated into an important causal factor in the light of high prevalence of susceptibility allele. As the systematic review result, the prevalence of susceptibility allele in controls group for XCCR1 Arg194Trp ranged from 2% in populations of Finland (50) and Saudi Arabia (39) to 57% in the population of Poland (37). For XRCC1 Arg280His the prevalence ranged between 3% in African-American population of USA (68) to 14% in population of India (28). For XRCC1 Arg399Gln, the prevalence ranged between 11% for African-American population of the US (83) to 57% for the population of Poland (37). For OGG1 Ser326Cys, the prevalence ranged between 21% for populations of Spain (73) and USA (27) to 68% for the population of Poland (87). The distribution of a susceptibility allele in a population is supposed to follow the Hardy-Weinberg law, unless a known population stratification or funder effect disturbs the HWE law (96). In studies of association between a susceptible allele and risk of cancer when there is no indication of population stratification or funder effect, the differential frequency of the susceptibility allele among the diseased compared to non-diseased provides the bases for association studies. Lack of HWE law in distribution of susceptible allele confounds any study assessing association between susceptibility allele and cancer risk (97, 98). In our systematic review, it was observed that HWE law was assessed for all the studies.
In association studies of susceptibility allele and risk of breast cancer, controlling other risk factors such as risk factors related to life style and hormonal play an important role. Many studies reviewed in this systematic review included control for known risk factors; in some studies, the analysis were done based on subgroup analysis and some studies used statistical modeling. Studies included in our analysis control extraneous risk factors either through subgroup analysis (grouping the study subjects into pre and post menopause) or by using multivariable models. Many of the studies were case-control studies some of which were population and clinical based studies. As population based case and controls are less prone to bias, our systematic review identifies many studies whose results can be used for further studies such as met-analysis.
In epidemiology, systematic reviews are done for different purposes. Our systematic review’s aim was to identify all studies that assessed the relationship between risk of breast cancer and susceptibility alleles of main base excision repair genes (XRCC1 and OGG1). The systematic approach makes sure that all studies are included and properly evaluated. The fact that our systematic review was able to use major databases (PubMed and Web of Sciences) indicates that we were able to identify all studies indexed in these two databases which cover a large part of English language indexed publications. The inclusion of two databases and manual search of the references of other systematic reviews and meta analyses have lower chances of missing major and important studies indicating high degrees of completeness.
The measure of association was another factor, which may be different among different populations. The difference depends on the frequency and magnitude of other modifying factors effecting and interacting with the BER genes. Our systematic review reveals that, the association between risk of breast cancer and XCRR1 and OGG1 susceptible allele depended on the model of inheritance and different model can show different effects on different population. Further studies are needed to assess the effect(s) of inheritance model on the risk of breast cancer.
7. Conclusions
Systematic search of major databases identify many studies addressing the relationship between breast cancer and susceptible alleles in the base excision repair genes and the fact that there are many variations in the magnitude of association depending on model of inheritance and the population of the study.
Timeframe of Search
The initial sensitive electronic search was performed from database commencement to 04 Sept 2014. The search updated in 22 April 2015. The details of strategy are shown in Appendix A1 (below table).
| Database | Search Strategy | N |
|---|---|---|
| (“Polymorphism”) AND (“Breast Neoplasms” OR “Breast Tumor” OR “Human Mammary Neoplasm” OR “Human Mammary Carcinoma” OR “Cancer of Breast” OR “Breast cancer” OR “Mammary Cancer” OR “Malignant Neoplasm of Breast” OR “Malignant Tumor of Breast” OR “Breast Carcinoma” OR “Cancer of the Breast” OR “Ductal carcinoma of breast” OR “Lobular Carcinoma of breast” OR “Medullary breast cancer” OR “Breast tumor” OR “In Situ breast cancer”) | 5345 | |
| (“Polymorphism”[ti]) AND (“Breast Neoplasms” [ti] OR “Breast Tumor” [ti] OR “Human Mammary Carcinoma” [ti] OR “Cancer of Breast”[ti] OR “Breast cancer”[ti] OR “Mammary Cancer”[ti] OR “Malignant Neoplasm of Breast”[ti] OR “Malignant Tumor of Breast”[ti] OR “Breast Carcinoma”[ti] OR “Cancer of the Breast”[ti] OR “Ductal carcinoma of breast”[ti] OR “Lobular Carcinoma of breast”[ti] OR “Medullary breast cancer”[ti] OR “Breast tumor”[ti] OR “In Situ breast cancer”[ti]) | 138 | |
| (Polymorphism | 1501 |
a Calculated OR for different strata, but data not shown.
| Study (year) | Source of Control | Population Ethnicity | Sample Size | Menopausal Status | HWE Reported |
|---|---|---|---|---|---|
| + | + | + | + | + | |
| + | - | + | + | + | |
| + | - | + | - | - | |
| + | + | + | + | + | |
| + | - | + | A | + | |
| + | + | + | + | + | |
| + | - | + | + | + | |
| - | - | + | - | + | |
| - | - | + | + | + | |
| + | - | + | - | + | |
| B | + | + | - | + | |
| + | A,C | + | + | + | |
| + | - | + | - | + | |
| + | + | + | + | + | |
| + | + | + | + | + | |
| + | D | + | + | + | |
| + | + | + | - | + | |
| + | - | + | - | + |
a Definitions: Year, year of publication; Source of control means clinic (or hospital) based or population based; population ethnicity means Caucasian, African-American and other ethnicity; Sample size means calculable sample size; Menopausal status means adjustment of menopausal status (post, pre or both) for final measure of association using methods such as adjustment in regression models, restriction, stratification etc; HWE (Hardy-Weinberg equilibrium) means calculated or reported HWE equilibrium.
b A, Adjusted, but no detail presented; B, Source of controls: volunteers; C, Matched controls, but no detail presented; D, Only mentioned to ethnicity without adjustment.
| Study (year) | Source of Control | Population Ethnicity | Sample Size | Menopausal Status | HWE Reported |
|---|---|---|---|---|---|
| + | + | + | A | + | |
| + | - | + | + | + | |
| + | B | + | C | + | |
| + | + | + | - | + | |
| + | + | + | - | + | |
| + | - | + | - | + | |
| + | - | + | - | + | |
| - | - | + | - | + | |
| + | - | + | + | + | |
| + | A | + | + | + | |
| + | BD | + | + | + | |
| + | + | + | - | + | |
| + | - | + | - | + | |
| + | + | + | C | + | |
| + | + | + | + | + | |
| + | + | + | + | + | |
| + | + | + | - | + | |
| + | - | + | + | + | |
| E | + | + | D | + | |
| E | + | + | - | + | |
| - | - | + | + | + | |
| + | D | + | C | + | |
| + | - | + | - | + | |
| E | + | + | - | + | |
| + | + | + | - | + | |
| + | - | + | - | + | |
| + | D | + | - | + | |
| + | + | + | - | + | |
| + | B | + | + | + | |
| + | - | + | A | + | |
| + | - | + | - | + | |
| - | - | + | - | + |
a Definitions:Year, year of publication; positive sign (+), mentioned in the articles; negative sign (-), not mentioned in the article; Source of control means clinic (or hospital) based or population based; Population ethnicity means Caucasian, African-American and other ethnicity; sample size means calculable sample size; Menopausal status means adjustment of menopausal status (post, pre or both) for final measure of association using methods such as adjustment inregression models, restriction, stratification etc; HWE (Hardy Weinberg equilibrium) means calculated or reported HWE equilibrium.
b A, Calculated OR for different strata, but data not shown; B, Only mentioned to ethnicity without adjustment; C, Adjusted, but no detail presented; D, Matched controls, but no detail presented; E, Source of controls: volunteers.
| Study (year) | Source of Control | Population Ethnicity | Sample Size | Menopausal Status | HWE Reported |
|---|---|---|---|---|---|
| + | - | + | - | + | |
| + | - | + | + | + | |
| + | + | + | A | + | |
| + | + | + | - | + | |
| + | + | + | + | + | |
| + | + | + | - | + | |
| B | + | + | - | + | |
| + | - | + | + | + | |
| + | C | + | D | + | |
| + | + | + | - | + |
a Definitions: Year, year of publication; Source of control means clinic (or hospital) based or population based; Population ethnicity means Caucasian, African-American and other ethnicity; Sample size means calculable sample size; Menopausal status means adjustment of menopausal status (post, pre or both) for final measure of association using methods such as adjustment in regression models, restriction, stratification etc; HWE (Hardy Weinberg equilibrium) means calculated or reported HWE equilibrium.
b A, Calculated OR for different strata, but data not shown; B, Source of controls: volunteers; C, Matched controls, but no detail presented; D, Adjusted, but no detail presented.
| Study (year) | Source of Control | Population Ethnicity | Sample Size | Menopausal Status | HWE Reported |
|---|---|---|---|---|---|
| + | + | + | A | + | |
| + | - | + | + | + | |
| + | B | + | C | + | |
| + | + | + | + | + | |
| + | + | + | - | + | |
| + | + | + | - | + | |
| + | - | + | - | + | |
| - | - | + | - | + | |
| + | B,C | + | - | + | |
| + | - | + | - | + | |
| + | - | + | - | - | |
| + | + | + | A | + | |
| + | - | + | + | + | |
| D | - | + | - | - | |
| + | BE | + | + | + | |
| + | A | + | + | + | |
| + | + | + | C | + | |
| + | B | + | - | + | |
| + | + | + | - | + | |
| + | - | + | - | + | |
| + | + | + | + | + | |
| + | + | + | + | + | |
| + | - | + | - | + | |
| + | + | + | + | + | |
| + | - | + | - | - | |
| D | + | + | - | + | |
| D | + | + | E | + | |
| D | + | + | - | + | |
| - | + | + | + | + | |
| + | + | + | - | + | |
| + | - | + | + | + | |
| - | - | + | + | + | |
| + | - | + | - | - | |
| + | + | + | - | + | |
| + | - | + | - | - | |
| + | - | + | - | + | |
| D | + | + | - | + | |
| D | - | + | - | + | |
| - | - | + | + | + | |
| + | E | + | C | + | |
| + | + | + | - | + | |
| + | C,E | + | + | + | |
| + | - | + | - | + | |
| + | E | + | - | + | |
| + | + | + | - | + | |
| + | - | + | - | + | |
| + | + | + | - | + | |
| + | + | + | + | + | |
| + | - | + | A | + | |
| + | + | + | - | + | |
| - | - | + | - | + |
a Year, year of publication; Source of control means clinic (or hospital) based or population based; Population ethnicity means Caucasian, African-American and other ethnicity; Sample size means calculable sample size; Menopausal status means adjustment of menopausal status (post, pre or both) for final measure of association using methods such as adjustment in regression models, restriction, stratification etc; HWE (Hardy Weinberg equilibrium) means calculated or reported HWE equilibrium.
b A, Calculated OR for different strata, but data not shown; B, Only mentioned to ethnicity without adjustment; C, Adjusted, but no detail presented; D, Source of controls: volunteers; E, Matched controls, but no detail presented.
| First Author | Date | Country | Ethnicity | Study Design | Control Source | Genotyping Methods | Analyzed Sample Size (Case Number) | Minor Allele Frequency (Case/Control) | Considered Confounders |
|---|---|---|---|---|---|---|---|---|---|
| 2001 | USA | African-Americans | population based | population based | multiplex PCR-RFLP and 59-exoncuclease (Taqman) | 327 (161) | (0.05/0.06) | age | |
| 2001 | USA | Whites | population based | population based | multiplex PCR-RFLP and 59-exoncuclease (Taqman) | 485 (251) | (0.05/0.06) | age | |
| 2002 | Korea | NS | hospital based | hospital based | PCR-RFLP | 410 (205) | (0.34/0.34) | breast feeding, family history of breast cancer and menopausal status at baseline | |
| 2003 | USA | Caucasian, Southern European, Scandinavian, Asians, Hispanics, and African Americans | population based | population based | pyrosequencing, RFLP, and the 5 nuclease assay (TaqMan) | 2367 (998) | (0.05/0.06) | age; menopausal status; postmenopausal hormone use; date of blood draw; time of blood draw; fasting status, body mass index at age 18 years, weight gain since age 18 years, age at menarche, age at menopause, parity/age at first birth, first-degree family history of breast cancer, history of benign breast disease, alcohol intake, during smoking, and duration of postmenopausal hormone use | |
| 2003 | France | NS | hospital based | population based | TaqMan technology or variant specific restriction enzyme digestion | 566 (254) | (0.07/0.06) | age | |
| 2003 | USA | Caucasian | hospital based | hospital based | PCR-RFLP | 512 (246) | (0.07/0.04) | age, family history, age at menarche, age at first live birth, and body mass index | |
| 2003 | USA | Caucasian | hospital based | hospital based | PCR-RFLP | 463 (162) | (0.07/0.06) | age | |
| 2004 | Turkey | NS | NS | NS | multiplex PCR | 284 (151) | (0.03/0.05) | - | |
| 2004 | Finland | NS | NS | population based | PCR-RFLP | 521 (223) | (0.03/0.02) | - | |
| 2005 | India | NS | hospital based | hospital based | PCR-RFLP and DNA sequencing techniques | 246 (123) | (0.21/0.12) | age and menopausal status | |
| 2005 | USA | white, African-American, Hispanic, Asian, other/unknown | population based | population based | Taqman | 970 (485) | NC | matched pairs, body mass index, age at menopause, age at menarche, alcohol intake, education, parity, age at first live birth, physical activity, history of breast cysts, family history of breast cancer in mother or sister, use of hormone replacement therapy, and smoking status | |
| 2005 | USA | White or non-White | population based | population based | fluorescence polarization method | 2177 (1067) | NC | age at menarche, parity, lactation, months of lactation, age at first birth, number of miscarriages, history of fertility problems, body mass index at reference, body mass index at age 20, first-degree family history of breast cancer, history of benign breast disease, menopausal status, oral contraceptive use, hormone replacement use, race, education, religion, and marital status | |
| 2006 | USA | Mostly Caucasian | population based | population based | patented fluorogenic method for nucleic acid analysis(Taqman or 5’nuclease assay) and PCR-RFLP | 615 (305) | (0.06/0.06) | family history | |
| 2006 | USA | African Americans | population based | Population based | the ABI 7700 sequence detection system or ‘‘Taqman’’ assay | 1456 (774) | (0.07/0.07) | offsets and age | |
| 2006 | USA | Whites | population based | population based | the ABI 7700 sequence detection system or ‘‘Taqman’’ assay | 2407 (1281) | (0.06/0.07) | offsets and age | |
| 2006 | USA | NS | population based | population based | PCR-RFLP, PCR-INVADER, or Sequenom | 626 (309) | (0.07/0.06) | age, family history of breast cancer among first degree relatives, physical activity, body mass index | |
| 2006 | USA | Caucasian | Population based | population based | TaqMan | 2833 (1580) | (0.06/0.07) | age and study site | |
| 2007 | Portugal | Caucasian Portuguese | hospital based | hospital based | PCR-RFLP | 693 (240) | (0.08/0.06) | age at diagnosis; alcohol consumption and smoking habits | |
| 2008 | Greece | Greek Cypriots | population based | volunteers | PCR-RFLP | 2261 (1097) | ( 0.08/0.08) | - | |
| 2008 | India | Caucasians of Indo-European | hospital based | volunteers | PCR-RFLP | 376 (151) | (0.06/0.14) | - | |
| 2008 | Thailand | NS | hospital based | hospital based | rapid capillary PCR followed by melting curve analysis using fluorescence labeled hybridization probes in a light cycler | 931 (506) | (0.30/0.31) | age, body mass index, age at menarche, family history of breast cancer, reproduction parameters, use of contraceptives, menopausal status, hazardous lifestyle, and education | |
| 2008 | USA | Caucasian | hospital based | Hospital based | mass ARRAY system | 735 (324) | (0.07/0.05) | age, family history of breast cancer, smoking history, age at menarche, age at first live birth and body mass index | |
| 2008 | USA | African-American | hospital based | hospital based | mass ARRAY system | 128(53) | (0.06/0.06) | age, family history of breast cancer, smoking history, age at menarche, age at first live birth and body mass index | |
| 2009 | Poland | NS | NS | NS | PCR-RFLP | 256 (150) | (0.53/0.57) | - | |
| 2010 | Taiwan | NS | hospital based | hospital based | real-time PCR with the TaqMan assay system | 934 (401) | (0.32/0.28) | age, age at first full-term pregnancy, cigarette smoking, body mass index, and family history | |
| 2010 | Italy | Caucasian | hospital based | volunteers | PCR-RFLP | 74 (43) | (0.08/0.04) | - | |
| 2010 | USA | NS | population based | population based | fluorescence polarization method | 559 (254) | NC | menopause and age and age at menarche | |
| 2011 | China | Han Chinese | hospital based | population based | PCR-RFLP | 1999 (995) | (0.30/0.27) | age | |
| 2013 | Saudi Arabia | NS | hospital based | hospital based | TaqMan SNP genotype analysis technique | 200(100) | (0.08/0.02) | - | |
| 2013 | Poland | NS | hospital based | population based | PCR-RFLP | 390 (185) | (0.14/0.08) | - | |
| 2014 | China | Han Chinese | hospital based | hospital based | polymerase chain reaction-ligase detection reaction (PCR-LDR) | 1239 (606) | (0.32/0.32) | age | |
| 2014 | USA | White and black | population based | population based | fluorescence polarization (FP) method | 1166 (598) | NC | age | |
| 2014 | Egypt | NS | hospital based | hospital based | PCR-RFLP | 175(100) | (0.40/0.20) | age, body mass index , parity, menopausal status, age at menarche, family history of breast cancer, smoking, or physical activity | |
| 2014 | Poland | NS | hospital based | hospital based | PCR-RFLP | 140 (70) | (0.49/0.50) | - | |
| 2015 | Mexico | NS | hospital based | NS | PCR-RFLP | 697 (345) | (0.16/0.15) | - |
a Definitions: Date, year of publication; NS, not state; NC, not calculable; USA, United State of America.
b Confounders in multivariate analysis.
| First Author | Date | Country | Ethnicity | Study Design | Control Source | Genotyping Methods | Analyzed Sample Size (Case Number) | Minor Allele Frequency (Case/Control) | Considered Confounders |
|---|---|---|---|---|---|---|---|---|---|
| 2003 | France | NS | hospital based | population based | TaqMan technology or variant specific restriction enzyme digestion | 566 (254) | (0.08/0.04) | age | |
| 2005 | India | NS | hospital based | hospital based | PCR-RFLP and DNA sequencing techniques | 246 (123) | (0.11/0.14) | age and menopausal status | |
| 2005 | Finland | Caucasian | hospital based | population based | PCR-RFLP | 959 (480) | (0.08/0.07) | age, age at menarche, age at first full term pregnancy, number of pregnancies, history of benign breast disease, first degree family history of breast cancer, waist-to-hip ratio, smoking and use of alcohol | |
| 2006 | USA | African Americans | population based | population based | the ABI 7700 sequence detection system or ‘‘Taqman’’ assay | 1446 (765) | (0.04/0.03) | offsets and age | |
| 2006 | USA | Whites | population based | population based | the ABI 7700 sequence detection system or ‘‘Taqman’’ assay | 2400 (1271) | (0.05/0.04) | offsets and age | |
| 2006 | USA | Caucasian | population based | population based | TaqMan | 2802 (1564) | (0.05/0.04) | age and study site | |
| 2008 | Greece | Greek Cypriots | population based | volunteers | PCR-RFLP | 2277 (1109) | ( 0.08/0.09) | - | |
| 2008 | Thailand | NS | hospital based | hospital based | rapid capillary PCR followed by melting curve analysis using fluorescence labeled hybridization probes in a light cycler | 932 (507) | (0.08/0.07) | age, body mass index, age at menarche, family history of breast cancer, reproduction parameters, use of contraceptives, menopausal status, hazardous lifestyle, and education | |
| 2008 | USA | Caucasian | hospital based | hospital based | mass ARRAY system | 732 (324) | (0.04/0.05) | age, family history of breast cancer, smoking history, age at menarche, age at first live birth and body mass index | |
| 2008 | USA | African-American | hospital based | hospital based | mass ARRAY system | 128 (53) | (0.03/0.04) | age, family history of breast cancer, smoking history, age at menarche, age at first live birth and body mass index | |
| 2010 | USA | NS | population based | population based | fluorescence polarization method | 599 (271) | NC | menopause and age and age at menarche | |
| 2011 | China | Han Chinese | hospital based | population based | PCR-RFLP | 1999 (995) | (0.09/0.10) | age |
a Definitions: Date, year of publication; NS, not state; NC, not calculable; USA, United State of America.
b Confounders in multivariate analysis.
| First Author | Date | Country | Ethnicity | Study Design | Control Source | Genotyping Methods | Analyzed Sample Size (Case Number) | Minor Allele Frequency (Case/Control) | Considered Confounders |
|---|---|---|---|---|---|---|---|---|---|
| 2001 | USA | African-Americans | population based | population based | multiplex PCR-RFLP and 59-exoncuclease (Taqman) | 519(253) | (0.19/0.14) | age | |
| 2001 | USA | Whites | population based | population based | multiplex PCR-RFLP and 59-exoncuclease (Taqman) | 767(386) | (0.35/0.36) | AGE | |
| 2002 | Korea | NS | hospital based | hospital based | PCR-RFLP | 410(205) | (0.35/0.31) | breast feeding, family history of breast cancer and menopausal status at baseline | |
| 2003 | USA | Caucasian, Southern European, Scandinavian, Asians, Hispanics, and African Americans | population based | population based | pyrosequencing, RFLP, and the 5 nuclease assay (TaqMan) | 2323(986) | (0.37/0.36) | age; menopausal status; postmenopausal hormone use; date of blood draw; time of blood draw; fasting status, body mass index at age 18 years, weight gain since age 18 years, age at menarche, age at menopause, parity/age at first birth, first-degree family history of breast cancer, history of benign breast disease, alcohol intake, during smoking, and duration of postmenopausal hormone use | |
| 2003 | France | NS | hospital based | population based | TaqMan technology or variant specific restriction enzyme digestion | 566(254) | (0.34/0.35) | age | |
| 2003 | China | Han Chinese | population based | population based | PCR-based RFLP | 2270(1088) | (0.28/0.27) | age, education, family history of breast cancer, menopausal status, age at menarche, age at menopause, body mass index, waist: hip ratio, and physical activity | |
| 2003 | USA | Caucasian | hospital based | hospital based | PCR-RFLP | 518(251) | (0.36/0.33) | age, family history, age at menarche, age at first live birth, and body mass index | |
| 2003 | USA | Caucasian | hospital based | hospital based | PCR-RFLP | 462(162) | (0.34/0.35) | age | |
| 2004 | Turkey | NS | NS | NS | multiplex PCR | 284(151) | (0.39/0.37) | - | |
| 2004 | Canada | Caucasian-Native or Caucasian-Asian or Caucasian-Black | population based | population based | matrix-assisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometry | 804(402) | (0.35/0.37) | ethnicity, age, and family history of breast cancer | |
| 2004 | Finland | NS | NS | population based | PCR-RFLP | 521(223) | (0.32/0.32) | - | |
| 2004 | Taiwan | NS | hospital based | hospital based | PCR-RFLP | 368(136) | (0.27/0.23) | - | |
| 2005 | India | NS | hospital based | hospital based | PCR-RFLP and DNA sequencing techniques | 246 (123) | (0.34/0.21) | age and menopausal status | |
| 2005 | Brazil | NS | hospital based | volunteers (among hospital personnel) | PCR-RFLP | 205(86) | (0.27/0.29) | - | |
| 2005 | Finland | Caucasian | hospital based | population based | PCR-RFLP | 957(479) | (0.30/0.27) | age, age at menarche, age at first full term pregnancy, number of pregnancies, history of benign breast disease, first degree family history of breast cancer, waist-to-hip ratio, smoking and use of alcohol. | |
| 2005 | USA | White, African-American, Hispanic, Asian, other/unknown | population based | population based | Taqman | 904(452) | (0.35/0.34) | matched pairs, body mass index, age at menopause, age at menarche, alcohol intake, education, parity, age at first live birth, physical activity, history of breast cysts, family history of breast cancer in mother or sister, use of hormone replacement therapy, and smoking status | |
| 2005 | USA | White or non-White | population based | population based | fluorescence polarization method | 2177(1067) | ( 0.36/0.35) | age at menarche, parity, lactation, months of lactation, age at first birth, number of miscarriages, history of fertility problems, body mass index at reference, body mass index at age 20, first-degree family history of breast cancer, history of benign breast disease, menopausal status, oral contraceptive use, hormone replacement use, race, education, religion, and marital status | |
| 2006 | USA | mostly Caucasian | population based | population based | patented fluorogenic method for nucleic acid analysis(Taqman or 5’nuclease assay) and PCR-RFLP | 615(305) | (0.38/0.37) | family history | |
| 2006 | USA | Caucasian, Black, Hispanic, Ashkenazi Jewish | hospital based | hospital based | multiplex primer extension PCR | 285(190) | (0.34/0.33) | - | |
| 2006 | USA | African Americans | population based | population based | the ABI 7700 Sequence Detection System or ‘‘Taqman’’ assay | 1437(761) | (0.16/0.14) | offsets and age | |
| 2006 | USA | Whites | population based | population based | the ABI 7700 sequence detection system or ‘‘Taqman’’ assay | 2366(1244) | (0.36/0.35) | offsets and age | |
| 2006 | USA | NS | population based | population based | PCR-RFLP, PCR-invader, or Sequenom | 515(193) | (0.50/0.36) | age, family history of breast cancer among first degree relatives, physical activity, body mass index | |
| 2006 | China | Han Chinese | hospital based | population based | PCR-RFLP | 941(302) | ( 0.25/0.26) | age, age at menarche and menopausal | |
| status | |||||||||
| 2006 | USA | Caucasian | population based | population based | TaqMan | 5626(3039) | (0.36/0.36) | age and study site | |
| 2006 | Poland | Caucasian | population based | population based | TaqMan | 4275(1985) | (0.37/0.35) | age and study site | |
| 2007 | Portugal | NS | hospital based | population based | PCR-RFLP | 917(256) | (0.34/0.39) | age | |
| 2007 | Portugal | Caucasian Portuguese | hospital based | hospital based | PCR-RFLP | 697(241) | (0.31/0.34) | age at diagnosis; alcohol consumption and smoking habits | |
| 2008 | USA | NS | hospital based | hospital based | PCR-RFLP | 88(40) | (0.52/0.53) | - | |
| 2008 | Slovakia | Caucasian | hospital based | volunteers | PCR with confronting two pair primers (PCR-CTPP) | 227(114) | (0.35/0.34) | - | |
| 2008 | Greece | Greek Cypriots | population based | volunteers | PCR-RFLP | 2283(1107) | ( 0.32/0.33) | - | |
| 2008 | India | Caucasians of Indo-Europeans | hospital based | volunteers | PCR-RFLP | 375(150) | (0.39/0.53) | - | |
| 2008 | Iran | Persian Muslims | hospital based | NS | PCR-RFLP | 373(186) | (0.36/0.32) | family history of breast cancer in first-degree relatives and cigarette smoking | |
| 2008 | Thailand | NS | hospital based | hospital based | rapid capillary PCR followed by melting curve analysis using fluorescence labeled hybridization probes in a Light Cycler | 931(507) | (0.27/0.23) | age, body mass index, age at menarche, family history of breast cancer, reproduction parameters, use of contraceptives, menopausal status, hazardous lifestyle, and education | |
| 2008 | USA | Caucasian | hospital based | hospital based | mass ARRAY system | 718(312) | (0.34/0.33) | age, family history of breast cancer, smoking history, age at menarche, age at first live birth and body mass index | |
| 2008 | USA | African-American | hospital based | hospital based | mass ARRAY system | 126(52) | (0.14/0.11) | age, family history of breast cancer, smoking history, age at menarche, age at first live birth and body mass index | |
| 2009 | Poland | NS | NS | NS | PCR-RFLP | 256(150) | (0.55/0.57) | - | |
| 2009 | India | NS | hospital based | hospital based | PCR-RFLP | 726(359) | (0.37/0.30) | age | |
| 2010 | Poland | NS | NS | NS | PCR-based fragment analyses using the CEQTM 8000 Genetic DNA Analysis System and/or ethidium bromide-stained agarose gels | 607(317) | (0.35/0.39) | index, year of birth and BRCA1 mutation | |
| 2010 | Poland | Caucasian | hospital based | hospital based | PCR-RFLP | 645(94) | (0.35/0.37) | - | |
| 2010 | Taiwan | NS | hospital based | hospital based | real-time PCR with the TaqMan assay system | 926(395) | (0.31/0.29) | age, age at first full-term pregnancy, cigarette smoking, body mass index, and family history | |
| 2010 | Poland | NS | hospital based | NS | PCR-RFLP | 440(220) | (0.44/0.34) | - | |
| 2010 | Brazil | NS | NS | volunteers | PCR-RFLP | 150(65) | (0.33/0.40) | - | |
| 2010 | Italy | Caucasian | hospital based | volunteers | PCR-RFLP | 74(43) | (0.53/0.32) | - | |
| 2010 | USA | NS | population based | population based | fluorescence polarization method | 555(254) | (0.30/0.34) | menopause and age and age at menarche | |
| 2011 | China | Han Chinese | hospital based | population based | PCR-RFLP | 1999(995) | (0.26/0.28) | age | |
| 2011 | USA | NS | hospital based | population based | matrix-assisted laser desorption ionization time-of-flightmass spectrometry | 2752(982) | (0.34/0.32) | age, education, race, smoking status, age at first birth, family history of breast cancer, history of benign breast disease, age at menarche and hormone therapy use | |
| 2012 | Egypt | NS | hospital based | hospital based | PCR-CTPP method | 200(100) | (0.37/0.30) | age | |
| 2013 | Saudi Arabia | NS | hospital based | hospital based | TaqMan SNP genotype analysis technique | 200(100) | (0.26/0.25) | - | |
| 2013 | Poland | NS | hospital based | population based | PCR-RFLP | 390(185) | (0.36/0.37) | - | |
| 2013 | Canada | Caucasian | population based | population based | sequenom iPLEX Gold platform | 7225(2720) | NC | body mass index | |
| 2014 | China | Han Chinese | hospital based | hospital based | polymerase chain reaction-ligase detection reaction (PCR-LDR) | 1239(606) | (0.30/0.25) | Age | |
| 2014 | China | Han Chinese | hospital based | hospital based | PCR-CTPP | 439(194) | (0.34/0.25) | age, age at menarche, pregnancy, menopausal status, body mass index, and family history of cancer | |
| 2014 | USA | White and Black | population based | population based | fluorescence polarization (FP) method | 1163(597) | NC | age | |
| 2014 | Egypt | NS | hospital based | hospital based | PCR-RFLP | 175(100) | (0.44/0.29) | age, body mass index, parity, menopausal status, age at menarche, family history of breast cancer, | |
| smoking, or physical activity | |||||||||
| 2014 | Russia | Caucasian | NS | hospital based | real-time PCR allelic discrimination with TaqMan probes | 1222(658) | (0.36/0.31) | age | |
| 2015 | Mexico | NS | hospital based | NS | PCR-RFLP | 686(345) | (0.29/0.23) | - |
a Definitions: Date, year of publication; NS, not state; NC, not calculable; USA, United State of America.
b Confounders in multivariate analysis.
| First Author | Date | Country | Ethnicity | Study Design | Control Source | Genotyping Methods | Analyzed Sample Size (Case Number) | Minor Allele Frequency (Case/Control) | Considered Confounders |
|---|---|---|---|---|---|---|---|---|---|
| 2003 | Korea | Korean | hospital based | hospital based | PCR-RFLP | 549 (265) | (0.56/0.55) | age, body mass index, family history of breast cancer, and parity factor | |
| 2003 | Japan | Japanese | hospital based | hospital based | PCR-CTPP | 389 (201) | (0.48/0.42) | age, body mass index , family history of breast cancer, and parity factor | |
| 2003 | Denmark | NS | population based | population based | real-time PCR | 869 (425) | (0.22/0.24) | - | |
| 2004 | Taiwan | NS | hospital based | hospital based | PCR-RFLP | 368 (136) | (0.58/0.60) | - | |
| 2006 | China | Han Chinese | population based | population based | fluorogenic 5´ nuclease assay with primers | 2269 (1102) | (0.58/0.58) | AGE, education level, menopausal status, and age at first live birth | |
| 2006 | USA | NS | population based | population based | template-directed dye-terminator incorporation assay with fluorescence polarization detection | 2134 (1041) | (0.22/0.22) | AGE and simultaneously adjusted for three SNPs | |
| 2006 | USA | Caucasian | population based | population based | TaqMan | 2815 (1571) | (0.21/0.21) | age and study site | |
| 2008 | Poland | NS | NS | NS | PCR-RFLP | 206 (100) | (0.51/0.57) | - | |
| 2008 | Thailand | NS | hospital based | hospital based | rapid capillary PCR followed by melting curve analysis using fluorescence labeled hybridization probes in a Light Cycler | 930 (506) | (0.55/0.50) | age, body mass index, age at menarche, family history of breast cancer, reproduction parameters, use of contraceptives, menopausal status, hazardous lifestyle, and education | |
| 2008 | Poland | NS | hospital based | NS | PCR-RFLP | 89 (41) | (0.52/0.68) | age | |
| 2010 | Taiwan | NS | hospital based | hospital based | real-time PCR with the TaqMan assay system | 934 (401) | (0.63/0.62) | age, age at first full-term pregnancy, cigarette smoking, body mass index, and family history | |
| 2010 | Italy | Caucasian | hospital based | volunteers | PCR-RFLP | 74 (43) | (0.31/0.29) | - | |
| 2011 | USA | NS | hospital based | population based | matrix-assisted laser desorption ionization time-of-flightmass spectrometry | 2941 (1054) | (0.22/0.22) | age, education, race, smoking status, age at first birth, family history of breast cancer, history of benign breast disease, age at menarche and hormone therapy use | |
| 2013 | Korea | NS | hospital based | population based | Illumina GoldenGate™method | 707 (346) | (0.47/0.49) | age, family history and body mass index | |
| 2013 | China | Han Chinese | hospital based | hospital based | PCR and high-resolution melting and sequencing | 1407 (630) | (0.60/0.56) | - | |
| 2014 | China | Han Chinese | hospital based | hospital based | PCR-CTPP | 439 (194) | (0.55/0.59) | age, age at menarche, pregnancy, menopausal status, body mass index, and family history of cancer | |
| 2014 | USA | White and black | population based | population based | fluorescence polarization (FP) method | 1135(580) | NC | age | |
| 2014 | Spain | Caucasian | hospital based | hospital based | SNPlex technology (based on oligation assay/polymerase chain reaction and capillary electrophoresis) | 2123(711) | (-/0.21) | age | |
| 2014 | Poland | NS | hospital based | hospital based | PCR-RFLP | 140 (70) | (0.49/0.50) | - |
b Confounders in multivariate analysis.