1. Background
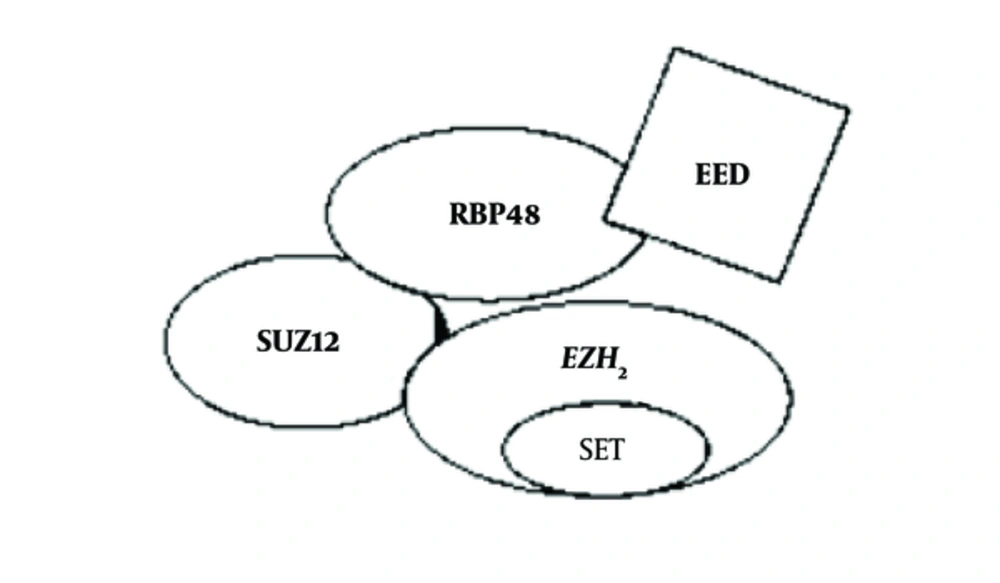
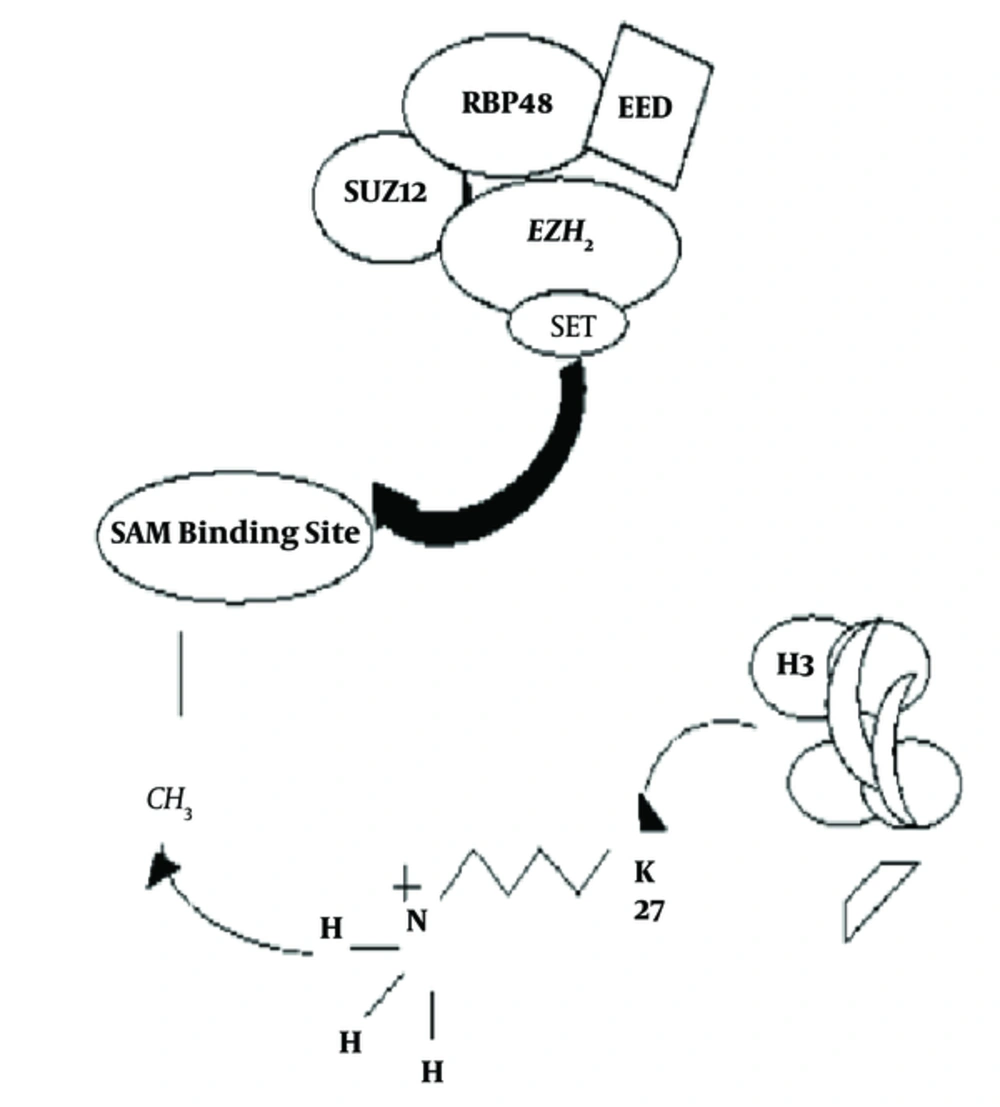
Polycomb group of proteins were initially identified as regulators that control the establishment of body segmentation, during embryogenesis, by silencing HOX genes, a subset of homeotic genes that are expressed in Drosophila. Later, it was found that they also act as epigenetic regulators, critical for multiple cellular functions as well as stem cell maintenance and differentiation (1). Polycomb group of proteins (PRC1 and PRC2) are conserved between Drosophila and human and are involved in gene silencing. PRC1 and PRC2, the 2 major polycomb repressive complexes, are known to control gene silencing through post-translational modifications of histone (2). The PRC2 protein complex contains EZH2, a histone methyltransferase that catalyzes trimethylation of histone H3 lysine 27 (H3K27me3) (3, 4). CBXs (Chromobox Homolog), PHC1 (Polyhomeotic Homolog 1), PHC2 (Polyhomeotic Homolog 2), PHC3 (Polyhomeotic Homolog 3), Ring1A (Really Interesting New Gene Domain of Polycomb Recessive Complex), Ring1B, BMI1 (B lymphoma Mo-MLV insertion region 1 homolog), and 6 PSC (Posterior Sex Comb Proteins) homologs comprise PRC1 complex. On the other hand SUZ12, EED, and RBP4 are part of the PRC2 complex. EZH2 is the catalytic subunit of the PRC2 protein complex, and its C-terminal SET domain exhibits the H3K27 methyltransferase function (Figure 1). It is seen that EZH2 has maximum catalytic activity for mono-methylation while reduced efficiency for the subsequent reactions (mono- to di- and di- to tri- methylations). The mechanism of methylation by EZH2 is majorly controlled by the S-Adenosyl Methionine (SAM) pocket located in the SET domain of the protein (Figure 2) (5). SET is a highly evolutionary conserved domain accountable for the catalytic activity of EZH2 (6). The SAM pocket has the sulfur atom from methionine, which acts as the methyl group donor. This forms an H-bond with the substrate and transfers the CH3 group to the amine nitrogen on H3K27. After the transfer of a single methyl group, the lone pair of electrons present at the amine N tends to orient away from the SAM pocket, rendering it lowly efficient for further methylations (5). EZH2 is currently considered a promising drug target, and multiple inhibitors of EZH2 have been developed, some of which are under clinical trials (6).
EZH2 is known to contribute towards cancer cell proliferation, migration, invasion, and metastasis by exhibiting cancer stem cell properties and tumor-initiating cell function (7-9). When EZH2 is overexpressed or mutated, a variety of cancers such as breast, prostate, lung, liver, colon, ovarian, bladder, leukemia, and lymphoma arise. The increased expression of EZH2 correlates with tumor malignancy and poor prognosis (10). In prostate cancers, the overexpression and amplification of EZH2 gene is hardly detected in early stage. Gene amplification of EZH2 is found in more than 50% of the hormone-refractory prostate cancers (11). The abnormal expression of EZH2 has been observed in breast epithelial cells, promoting tumorigenesis (12). Patients with myeloid malignancies such as Myelodysplastic syndrome and myeloproliferative neoplasm are seen to have inactivating mutation of EZH2 with very less rate of survival (13, 14). Other than myeloid malignancies, in 25% of T-cell leukemia, loss-of-function mutations and deletions of EZH2 and SUZ12 genes are found (15). The conditional deletion of EZH2 in bone marrow cells resulting in T-cell leukemia can also be considered one of the indicators of tumor suppressing properties of EZH2. Impaired pancreatic regeneration and acceleration of K-Ras induced neoplasias also result from conditional deletion of EZH2 in pancreatic epithelium (16, 17). Thus, the paradoxical role of EZH2 makes it an interesting target for research since the overall rate of survival for EZH2 mutations is poor.
1.1. Mode of Action of EZH2
The catalytic subunit of human PRC2 subunit - EZH2, acts by tri-methylating Lysine at the 27th position of histone-3 (H3K27) protein on the DNA. EZH2 is majorly involved in chromatin condensation and gene silencing. The major contribution is that its over-expression leads to silencing the tumor-suppressor genes (TSG) through the increased levels of histone methylation in the promoter regions of TSGs. EZH2 is mainly expressed during embryonic stages of development, having very depressed levels of expression during adult stages (18, 19). EZH2 level is regulated by JAK2/βTrCP (Janus Kinase 2/β-transducing-repeat-containing protein) complex. βTrCP (β-transducing-repeat-containing protein) marks EZH2 for ubiquitination following, which is phosphorylated by JAK2 at the Y641 residue located in the SET domain (6). This phosphorylation by JAK2 subsequently allows βTrCP mediated degradation of EZH2. Hence, this process maintains the normal EZH2 levels in a normal adult cell (18, 19).
1.2. Missense Mutations in EZH2
Non-synonymous single nucleotide polymorphisms (nsSNPs) are coding variants. It introduces amino acid changes in their corresponding proteins. Since nsSNPs can affect protein function, it is believed that they have a largest impact on human health and contribute towards many disease conditions. Hence, it is essential to distinguish the nsSNPs affecting protein function from those that are functionally neutral. In the present study, we attempted to retrieve deleterious mutations (SNPs) in various regions of EZH2 on the basis of their incidence of occurrence in cancer from COSMIC database and predict potential biomarkers for diagnostic purpose, using various online tools.
2. Methods
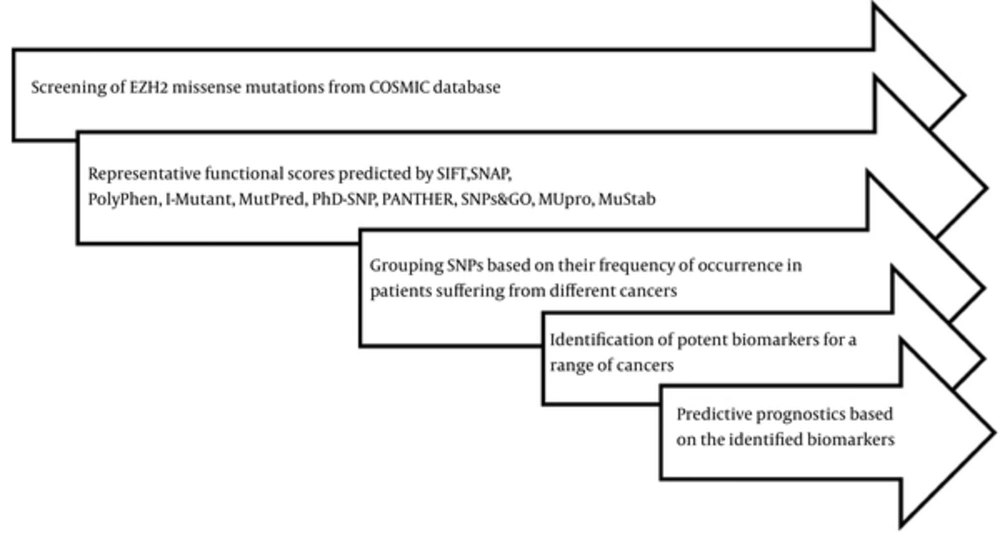
Specific EZH2 missense mutations were mined from the COSMIC database, and certain online bioinformatics tools such as SIFT, SNAP, PolyPhen 2.0, I-Mutant, MutPred, PhD-SNP, PANTHER, MUpro, MuStab, and SNPs&GO were applied to predict their representative functional scores. These tools, based on validated algorithms, helped detect the severity of mutations. In addition, these tools also helped analyze some structural or functional alterations in the coded protein. The SNPs were grouped based on their frequency of occurrence in various leukemia and lymphomas, which in turn assisted us to identify potent biomarkers for the cancer types taken up in the present study. Further prognostic effects of these SNPs were predicted and discussed for targeted treatments and diagnosis. Figure 3 describes the flow of work.
2.1. Tools Used for Various Analyses of Selected ns-SNPs
PolyPhen 2.0 searches for 3D protein structure, numerous homologue sequence alignment, and amino acid information in several protein structure databases. The Position specific independent count (PSIC) scores are calculated for each of 2 variants and the difference is calculated. The more the PSIC score is different for 2 variants, the more the functional impact of particular amino acids will be (http://genetics.bwh.harvard.edu/pph2/) (20). In PANTHER, Pdel denotes the probability of a variant, causing deleterious effect on a protein in such a way that a subPSEC score of -3 resemble to Pdel of 0.5. An evolutionary score is computed here and the method predicts deleterious or neutral effects with a probability score (http://PANTHERdb.org/) (21). PhD-SNP predicts deleterious SNPs for human based on Support Vector Machine (SVM). The output consists of the number of mutated position in protein sequence, the wild type residue and the novel or mutated residue and the mutation is predicted to be as diseased or neutral (http://snps.biofold.org/phd-snp/phd-snp.html) (22). SNPs&GO includes GO annotations as features in a SVM model to predict whether a SNP is a neutral one or associated with disease (http://snps.biofold.org/snps-and-go//snps-and-go.html) (23). I-Mutant3.0 is a support vector machine (SVM)-based tool that automatically predicts the protein stability changes upon single point mutations. I-Mutant3.0 predictions are performed starting either from the protein structure or from the protein sequence. I-Mutant3.0 programs can be used to predict the sign of the stability change upon mutation or as estimation of regression to predict changes in free energy (http://gpcr.biocomp.unibo.it/cgi/predictors/I-Mutant3.0/I-Mutant3.0.cgi) (24). MUpro (http://mupro.proteomics.ics.uci.edu/) uses both SVM and Neural Networks programs. The sequence-based version of the program was used here. The SVM method was run, using the default parameters. The output of the program deals with the sign of the energy change (25). SIFT determines the probability of substitution being tolerated in a given position (http://sift.jcvi.org/) (26). SNAP facilitates interpretation and comparison of genome-wide association study results (https://www.broadinstitute.org/mpg/snap/) (27). MuStab (http://bioinfo.ggc.org/MuStab/) is designed to predict protein stability that is changed due to amino acid substitution. An amino acid sequence in FASTA format needs to be entered by the user along with specific substitution, the pH condition, and temperature (28). MutPred is used to predict diseased or neutral SNPs. The features used refer to a probability of loss or gain of function regarding several functional and structural properties of the encoded protein (http://mutpred.mutdb.org/) (29).
3. Results
A set of missense mutations in various leukemia and lymphoma were mined from COSMIC database. These mutations were analyzed, using different tools as described earlier. Some of these mutations such as S651P, I669M, S651L, R646H, N631K, N649K, R615K, D620G, F626L, A677G, D124H, D136G, Y641N, Y641F, Y641S, and Y641C were predicted to have a high negative impact on the disease possibly contributing towards the accelerated progression of lymphatic and myeloid cancers. The most potent biomarkers were identified based on their representative functional scores as shown in the table (Tables 1 and 2) below. A few mutations mined from the COSMIC database were dealt separately with different online tools SIFT, SNAP, MUpro, and MuStab (Table 3) to understand their detrimental effects on the cancer progression. The results obtained from MutPred (Table 4) that focus on structural and functional alteration of proteins are tabulated below. Most of the protein analysis tools required the amino acid sequence of EZH2 protein from UniProt database. In the present study, we used UniProt ID - Q15910-1 and Q15910-3, since all mutations were found to be falling in the regions of isoform 1 or 3 of EZH2. As shown in the earlier results, all damaging mutations, which have a deleterious role in various hematologic cancers, are summarized in Table 5. These biomarkers may have the potential to be targeted in diagnostics and/or therapeutics. All these dynamic changes lead to altered substrate binding, causing altered protein properties with change in hydrophobicity and free energy.
| Mutation | PolyPhen 2.0 | PANTHER | PhD-SNP | SNPs&GO | I-Mutant ΔΔG, Kcal/mol | MutPred |
|---|---|---|---|---|---|---|
| S651P | 1.000,PrD | 0.752, D | 7, D | 0.763, D | -0.54 | 0.929 |
| I669M | 0.988,PrD | 0.641, D | 3, D | 0.516, D | -1.4 | 0.818 |
| S651L | 0.997,PrD | 0.767, D | 6, D | 0.629, D | -0.2 | 0.933 |
| R646H | 1.000,PrD | 0.643, D | 4, D | 0.550, D | -1.5 | 0.902 |
| N631K | 1.000,PrD | 0.515, D | 3, D | 0.564, D | -0.98 | 0.596 |
| R615K | 1.000,PrD | 0.568, D | 5, D | 0.630, D | -0.93 | 0.862 |
| N649K | 1.000,PrD | 0.752, D | 8, D | 0.841, D | -0.61 | 0.943 |
| D620G | 1.000,PrD | 0.769, D | 6, D | 0.793, D | -1.61 | 0.534 |
| F626L | 1.000,PrD | 0.587, D | 2, D | 0.568, D | -1.79 | 0.783 |
| D124H | 0.988,PrD | 0.587,D | 1, D | 0.645, D | -2.98 | 0.554 |
| D136G | 0.929,PoD | 0.427,N | 5, D | 0.607, D | -4.65 | 0.462 |
| Y641C | 0.180,B | 0.936,D | 0, D | 0.749, D | -1.46 | 0.93 |
| Y641F | 0.964,PrD | 0.507,D | 4, D | 0.653, D | -1.13 | 0.935 |
| Y641N | 0.979,PrD | 0.868,D | 1, D | 0.682, D | -1.25 | 0.934 |
| A677G | 1.000,PrD | 0.572D | 2, N | 0.583, D | -1.3 | NE |
| Y641S | 0.543,PoD | 0.844,D | 1, D | 0.768, D | -1.44 | 0.92 |
| Mutation | PolyPhen 2.0 | PANTHER | PhD-SNP | SNPs&GO | I-Mutant ΔΔG, Kcal/mol | MutPred |
|---|---|---|---|---|---|---|
| G5W | 1.000,PrD | NE | 7,N | 0.323,N | 0.56 | 0.21 |
| T93S | 0.000,B | 0.072,N | 8,N | 0.017,N | -1.86 | 0.313 |
| F120Q | 1.000,PrD | 0.512,D | 6,N | 0.545,D | -0.51 | 0.734 |
| I150R | 0.726,PoD | 0.265,N | 1,D | 0.615,D | -2.36 | 0.568 |
| N152M | 0.998,PrD | 0.488,N | 2,N | 0.450,N | -1.33 | 0.374 |
| R288Q | 0.893,PoD | 0.379,N | 5,N | 0.564,D | -1.11 | 0.42 |
| Q465Y | 0.998,PrD | 0.555,D | 9,N | 0.421,N | 0.32 | 0.412 |
| A482R | 0.168,B | 0.113,N | 7,N | 0.053,N | 0.77 | 0.293 |
| K495T | 1.000,PrD | 0.439,N | 6,N | 1.50,N | -0.6 | 0.392 |
| D581V | 1.000,PrD | 0.808,D | 7,N | 0.552,D | -1.45 | 0.559 |
| I626M | 0.997,PrD | 0.412,D | 4,N | 0.233,N | 0.55 | 0.624 |
| Y641H | 0.997,PrD | 0.799,D | 0,N | 0.647,D | -1.58 | 0.92 |
| V657H | 1.000,PrD | 0.917,D | 8,N | 0.528,D | -1.61 | 0.593 |
| K660V | 1.000,PrD | 0.694,D | 8,N | 0.562,D | -1.7 | 0.666 |
| N668V | 1.000,PrD | 0.686,D | 4,N | 0.378,N | -0.51 | 0.613 |
| E701K | 0.749, PoD | NE | 4, D | 0.500,N | -0.48 | 0.49 |
| P533L | 1.000,PrD | 0.695, D | 7, N | 0.200,N | -0.44 | 0.48 |
| R303Q | 0.003,B | 0.177, N | 5, N | 0.067,N | -0.64 | 0.249 |
| R16Q | 0.898, PoD | 0.234, N | 1, D | 0.212,N | -1.09 | 0.192 |
| H240Y | 1.000, PrD | 0.481, N | 5, D | 0.553,D | 0.36 | 0.447 |
| R640H | 0.862,PoD | 0.696, D | 3, D | 0.451,N | -1.58 | 0.612 |
| N516K | 0.990,PrD | 0.447, N | 2, D | 0.436,N | -0.32 | 0.259 |
| T222N | 1.000,PrD | 0.475, N | 2, D | 0.414 ,N | -0.85 | 0.163 |
| T639I | 1.000,PrD | 0.353, N | 7, D | 0.376 ,N | -1.15 | 0.766 |
| Mutation | SIFT | SNAP | MUpro | MuStab |
|---|---|---|---|---|
| D124H | 0.00, P | NoN, RI = 3, EA 78% | - | - |
| D136G | 0.00, P | NoN, RI = 2, EA 70% | - | - |
| Y641C | 0.00, P | NoN, RI = 6, EA 93% | - | - |
| S651P | - | - | -0.059 | 83.57, DS |
| A677G | 0.02, P | N, RI = 0, EA 53% | - | - |
| Y641S | 0.00, P | NoN, RI = 5, EA 87% | - | - |
| I669M | - | - | -0.193 | 89.64, DS |
| S651L | - | - | 0.208 | 77.68, DS |
| Y641F | 0.00, P | NoN, RI = 5, EA 87% | - | - |
| Y641N | 0.00, P | NoN, RI = 6, EA 93% | - | - |
| R646H | - | - | -1 | 83.93, DS |
| N631K | - | - | -0.269 | 88.93, DS |
| R615K | - | - | -0.355 | 86.07, DS |
| N649K | - | - | -1 | 79.64, DS |
| D620G | - | - | -0.979 | 84.11, DS |
| F626L | - | - | -0.277 | 92.5, DS |
| Mutation | Secondary Structure | Catalytic Residue | Methylation Sites | Ubiquitination Sites | MoRF Binding | Solvent Accessibility |
|---|---|---|---|---|---|---|
| S651P | Loss of sheet | - | - | - | Loss | Gain |
| S651L | Gain of Loop | Loss | - | - | Gain | - |
| R646H | Gain of Loop | - | Gain | Gain | Loss | - |
| I669M | Gain of sheet | Gain | - | Loss | Gain | - |
| N631K | - | - | Gain | Gain | Gain | Gain |
| R615K | - | - | Gain | Gain | - | Gain |
| N649K | - | Loss | Gain | - | Gain | - |
| D620G | - | Gain | Gain | Loss | - | - |
| F626L | Gain of sheet | Loss | Loss | Loss | - | - |
| Y641C | Gain of sheet | - | - | - | - | - |
| Y641F | Gain of sheet | - | - | - | - | - |
| Y641S | Gain of sheet | - | - | - | - | Gain |
| Y641N | Gain of sheet | - | - | - | - | Gain |
| A677G | Gain of loop | - | - | Gain | Loss | - |
| Mutation | Disease Outcome |
|---|---|
| Y641N, Y641F, Y641S, Y641C | Diffuse Large B-Cell Lymphoma & Follicular Lymphoma |
| A677G | Diffuse Large B-Cell Lymphoma |
| S651P, S651L, R646H | Acute Myeloid Leukemia |
| I669M | T cell-Acute Lymphoblastic Leukemia |
| N631K | Acute Lymphoblastic Leukemia |
| R615K, N649K, D620G, F626L | Chronic Myeloid Leukemia |
The deleterious mutations mentioned in Table 1 were also run through online tools SIFT, SNAP, MUpro, and MuStab. The mutations D124H, D136G, Y641C, A677G, Y641S, Y641F, Y641N predicted to be damaging by SIFT tool as per the functionality of these proteins since the scores of these mutations are less than 0.05 (26). The same sets of mutations were predicted to be non-neutral by SNAP tool (27). The other set of deleterious mutations S651P, I669M, S651L, R646H, N631K, N649K D620G, and F626L were analyzed, using MUpro and MuStab. MuStab showed a decreased stability for all these missense substitution with a prediction confidence of 83.57% (S651P), 89.64% (I669M), 77.68% (S651L), 83.93% (R646H), 88.93% (N631K), 86.07% (R615K), 79.64% (N649K), 84.11% (D620G), 92.5% (F626L) respectively (28). MUpro, with the help of support vector machine, assisted us to detect the decrease in protein stability with a confidence score of -0.059 (S651P), -0.193 (I669M), -1 (R646H and N649K), -0.269 (N631K), -0.355 (R615K), -0.979 (D620G), -0.277 (F626L) respectively and an increase in protein stability with a score of 0.208 in case of S651L substitution (25). The results are shown below.
The MutPred scores of all damaging missense substitutions are enlisted in Table 1. The following table (Table 4) focuses on some of the top features obtained from MutPred tool, which was observed for all these types of mutations such as alteration in secondary structure, change in catalytic residue, methylation and ubiquitination sites, MoRF binding, and solvent accessibility. For example, there was a confident hypothesis on loss of sheet (P = 0.0126) for mutation S651P, gain of loop (P = 0.0312) for S651L substitution, loss of sheet (P = 0.0126) and loss of MoRF binding (P = 0.0212) for substitution R646H, gain of sheet (P = 0.039) for I669M, gain of ubiquitination (P = 0.0369, P = 0.023) and methylation (P = 0.0379, P = 0.0183) for N631K and R615K, gain of relevant solvent accessibility (P = 0.0479) for R615K and gain of MoRF binding (P = 0.0256) and methylation (P = 0.029) for substitution N649K (29). For A677G substitution, a very confident hypothesis predicted was changes in secondary structures by means of gain of loop (P = 0.0312) and loss of sheet (P = 0.007). For Y641S substitution, the results in MutPred showed gain of phosphorylation (P = 0.1352) at Y641 residue and a gain of disorder with a prediction score of 0.0096. But, in Y641N, Y641F, and Y641C substitution, there is a loss of phosphorylation at Y641 residue. In this way, all these features that altered upon mutation exhibited deleterious effects.
Table 5 consists of all the deleterious missense substitutions that were found in various cancers dominantly Leukemia and Lymphomas screened from COSMIC database. These mutations hence can be considered as an important marker in the field of diagnosis and therapeutics.
4. Discussion
EZH2 is seen to regulate gene expression that helps control self-renewal of cells or maintain a balance during cellular differentiation. In the present study, we have used several in silico tools in order to predict the mutational effects of certain missense mutations mined from the COSMIC database for cancers in the EZH2 cluster. Loss-of-function and gain-of-function mutations in various regions of the genome result in modification of the structural framework of the coded proteins, making them highly unstable and rendering them inactive. The overall scores obtained in various missense mutations of EZH2 focuses on H-bond changes, hydrophobicity, altered stability, effect on secondary structures like loss or gain of sheet or loop, solvent accessibility, change in methylation sites, ubiquitination sites, catalytic residues, and alterations of MoRFs (Molecular Recognition Features). There are 3 basic types of MoRFs: α-MoRFs, which form α-helices, β-MoRFs that helps form β-strands, and i-MoRFs that forms an irregular secondary structure when bound (30).
In the current study, some mutations (Table 1) were found to be deleterious by various online bioinformatics tools. Hydrophobic effect is one of the major factors that drive a protein towards collapse and misfolding. If there is any increase or decrease in hydrophobicity upon mutation, it will disrupt the protein structure and function (31). In these missense mutations (S651P, S651L, I669M, R646H, R615K, F626L, D620G, N631K, N649K, A677G, Y641N, Y641F, Y641S, Y641C), there is a change in hydrophobicity. There is also a possibility for these mutant proteins to fold properly but being less stable, or executing a stable confirmation and, thus, making the protein dysfunctional (32).
Gain or loss of sheet (S651P, I669M, F626L, Y641N, Y641F, Y641S, and Y641C) contributes towards early events in disease onset and progression, whereas gain or loss of loop (S651L, R646H, and A677G) increases the surface area, not necessarily causing immediate deleterious effects (32, 33).
Altered methylation sites hold an important outcome resulting into tumor suppressor gene silencing and, thus, increasing cell proliferation. In this study, most of the mutations altered the methylation sites at residues R646H, N631K, R615K, N649K, D620G, and F626L respectively.
Variation in the ubiquitination sites (R646H, N631K, R615K, D620G, and F626L) affects the degradation via the proteasome pathway, alters cellular location of proteins, changes the protein activity, and alters protein interactions.
The significance of solvent accessibility is determined by the accessible surface area which predicts protein stability, as hydrophobic transfer energy is directly a measure of residue-wise solvent accessible surface (32). An altered solvent accessibility was observed for substitutions S651P, N631K and R615K, Y641S, Y641N.
In myeloid leukemia, EZH2 mutation is seen to affect the exons and even introns in some cases. The mutations discussed above were predicted to be inactivating leading to either formation of a truncated protein with deletion in the SET domain or a loss of amino acid essential for protein activity. Such missense mutations can also be associated with either loss of one copy of EZH2 gene or to a loss of heterozygosity (13, 14).
4.1. Discussion of Individual Potentially Damaging Biomarkers
4.1.1. S651P and S651L
Serine can reside both within the interior of a protein or on the protein surface. The serine side chain hydroxyl oxygen forms a hydrogen bond with the protein backbone, thereby it effectively mimicks Proline, which is an amino acid. On the other hand, being hydrophobic, Leucine buries itself in protein hydrophobic cores (33). When there is a substitution form Serine to Proline (S651P) or form Serine to Leucine (S651L), the hydrophobicity drops down from -0.8 to -1.6 or increased from -0.8 to 3.8 (kdHydrophobicity), respectively (31, 34, 35). Any disruption in hydrophobic interaction destabilizes the protein structure. The ΔΔG value from I-Mutant predicted to be -0.54 Kcal/mol and -0.20 Kcal/mol, which indicates a large decrease and increase in protein stability, respectively, for S651P and S651L, thereby causing a damaging effect. Mutations that affect and/or introduce proline considered to be significant. In S651P substitution, serine is substituted by proline, causing a different stress in polypeptide backbone and a steric clash in neighboring residue side chain due to the introduction of a pyrolidine ring in alpha helices and beta strands (32). A study by Dolnik et al. investigated mutations in the histone methyltransferase gene EZH2. The results obtained from this study showed that mutations in hematopoietic malignancies affected EZH2 gene in a non-persistent manner; thus, these mutations have a potential pathogenic role in treatment of cancer (36).
4.1.2. D620G
Aspartate is substituted by glutamate or other polar amino acids. Glycine contains hydrogen as its side chain, which provides it conformational flexibility (33). When there is a substitution form Aspartate to Glycine (D620G), the hydrophobicity increases from -3.5 to -0.4 (kdHydrophobicity). The ΔΔG value from I-Mutant predicted to be as -1.61 Kcal/mol (31, 34, 35). The top features of this substitution were gain of methylation at K621 residue, loss of ubiquitination at K617 residue, loss of phosphorylation at Y622 residue, and gain of a catalytic residue at Y619 residue (MutPred), which made this mutation to be a damaging one. This particular substitution is also considered to be significant, because an introduction of glycine can creates a hollow hydrophobic part resulting in protein destabilization (32).
4.1.3. I669M
Isoleucine can be substituted by hydrophobic, aliphatic amino acids. It prefers to be buried in protein hydrophobic cores because of its hydrophobicity. Methionine contains a sulphur atom, which is connected to a methyl group (33). A substitution form Isoleucine to Methionine (I669M) drops down the hydrophobicity from 4.5 to 1.9 (kdHydrophobicity). The free energy change from I-Mutant predicted to be as -1.40 Kcal/mol, which indicates a decrease in protein stability (31, 34, 35). Overall, this mutation has the ability to trigger T cell Acute Lymphoblastic Leukemia (Table 5).
4.1.4. R646H and R615K
Arginine functions in forming salt-bridges, pairing with negatively charged amino acids to create stabilized hydrogen bonds. Histidine is generally considered to be a polar amino acid, but it is significant with respect to its chemical properties (33). A substitution form arginine to histidine (R646H) changes the hydrophobicity from -4.5 to -3.2 (kdHydrophobicity). The ΔΔG value from I-Mutant predicted to be as -1.50Kcal/mol, thereby indicating to be deleterious with a confidence score -1(MUpro).
Lysine has a positively charged amino group on its side chain that functions in the formation of hydrogen bonds with negatively-charged non-protein atoms (33). A substitution form arginine to Lysine (R615K) changes the hydrophobicity from -4.5 to -3.9 (kdHydrophobicity). Any disruption in hydrophobic interaction destabilizes the protein structure. The ΔΔG value from I-Mutant predicted to be as -0.93Kcal/mol, which indicates a decrease in protein stability (31, 34, 35). Finally, these 2 missense substitution can serve as one of the prominent reasons for AML and CML, respectively (Table 5).
4.1.5. N631K and N649K
Asparagine generally prefers to be on the surface of proteins. It is involved in protein active sites or protein binding sites. Lysine has a positively charged amino group on its side chain, which forms hydrogen bonds with negatively-charged non-protein atoms (33). When there is a substitution form asparagine to lysine (N631K), the hydrophobicity changes from -3.5 to -3.9 (kdHydrophobicity) (31, 34, 35). The ΔΔG value from I-Mutant predicted to be as -0.98Kcal/mol, which indicates a decrease in protein stability. There is a possibility for these 2 mutations to occur in patients with ALL and CML (Table 5).
4.1.6. F626L
Leucine is hydrophobic in nature. It prefers to be covered up in protein hydrophobic core and to be with alpha helices. Phenylalanine particularly favors to exchange with tyrosine that contains hydroxyl group in place of ortho hydrogen in benzene ring (33). When there is a substitution form phenylalanine to Leucine (F626L), the hydrophobicity increases from 2.8 to 3.8 (kdHydrophobicity). The ΔΔG value from I-Mutant predicted to be as -1.79 Kcal/mol. Thus, the amino acid substitution has shown to decrease the protein stability, therefore, indicating towards a deleterious effect (31, 34, 35). Due to disruption in molecular mechanism, there is a loss of catalytic residue at F626 position, loss of methylation, and ubiquitination at K621 residue according to MutPred scoring. This mutation can also be found in patients with CML (Table 5) and, hence, it can serve as a diagnostic marker.
4.1.7. Y641X
Y641X (X = N, F, S, C) residue lies in the SET domain of EZH2. Substitution of tyrosine from the 641 position by other residues confer gain of function, thus, rendering to EZH2’s hypermethylating activity and, hence, gene silencing (37). The hydrophobicity drops down in Y641N substitution (-1.3 to -3.5), but increases in substitutions Y641F and Y641C (31). Normally EZH2 displays maximum catalytic activity for monomethylation of H3K27 and weaker ability for subsequent reactions. However, in Y641X mutants, it exhibits limited ability to mono-methylate but an enhanced catalytic efficiency for subsequent reactions. Y641X mutants work in conjunction with the wild type EZH2 to elevate the levels of H3K27me3 (38). All the somatic mutations targeting Y641 residue result in a greater stability and an increased half-life of EZH2 protein. This residue plays a very important role in Jak2/ βTrCP mediated degradation of EZH2 (39, 40).
4.1.8. A677G
A677 residue is also located in the SET domain of EZH2 (41). Like Y641X mutations, A677G mutation also confers gain of function, thus, hyper-methylating activity of EZH2 in DLBCL (41). The hydrophobicity changes from -4.5 to -0.4 (31). Substitution of Alanine to other Glycine leads to an increased activity with H3k27me2 substrates similar to Y641X mutations (42). The protein functionality is maintained by a combination of buried hydrophobic surfaces and interference with H-bonding of the protein with the surrounding solvent (41). However, it also retains H3K27me1 activity like wild type EZH2 .This eventually allows efficient utilisation of all 3 methylation substrates (me0, me1 & me2) (42).
5. Conclusions
EZH2 is a crucial element in cancer progression. Targeting the mutations in this gene can be a very potent solution for formulating anti-cancer treatments. Also, several specific mutations can act as potent biomarkers for different stages of cancer manifestation and, thus, have a role in diagnostics. A large set of experimental data show that the oncogenic role of EZH2 mainly depends on its ability to repress gene expression programs via H3K27 methylation and chromatin compaction. EZH2 is frequently overexpressed in multiple cancers and is associated with poor prognosis. Therefore, EZH2 may serve as a valuable prognostic marker. Overexpression of EZH2 is mainly found in solid tumors. Activating mutations are found in B-cell lymphomas. In myeloid disorders, EZH2 behaves like a tumor suppressor gene. EZH2 could be involved in cancer through multiple mechanisms and it could also be regulated by different pathways that depend on cellular context and cancer type. In this study, missense sub mutations selected from the COSMIC database are presented along with their corresponding functional scores in order to determine the potent biomarkers that could be useful for diagnosis and therapeutic study. In future, additional studies will be required to establish effective combination treatment strategies and identify appropriate biomarkers in various cancer types to predict sensitivity to EZH2 inhibitors. The upstream regulators of EZH2, if identified, may lead to effective therapeutic strategies for various cancers.