1. Background
Breast cancer is one of the leading causes of death in women worldwide (1), and it accounts for 23% of cancers among Iranian women (2). The incidence and progression of breast cancer are complicated processes led by various genetic and environmental factors (3-5). Despite great efforts in the area, the molecular mechanisms involved in the tumorogenesis sand progression of breast cancer remain unknown.
Recently, a new biological system has been suggested for classification of invasive breast carcinomas (6). This intrinsic system works based on the expression of Antigen Ki-67, human epidermal growth factor receptor 2 (HER2), estrogen receptor (ER), and progesterone receptor (PR) (7); accordingly, five distinct molecular subtypes are defined: Luminal A, Luminal B (HER2-), Luminal B (HER2+), HER2-rich, and basal-like subtypes (6). Luminal A accounts for 50-60% of breast carcinomas and is pathologically ER +, PR + and HER-, and very low Ki67 expression level. The prevalence of Luminal B is 15-20%, with expression of markers similar to those of Luminal A (8). Luminal B subtype has more aggressive phenotype (8) and involvements of lymph nodes in the Luminal B (HER2-) is higher compared with the Luminal A (9).
Since the structure and function of breast cancer cells differ from those of the normal cells, the protein content is different as well (10). In addition, the secretory proteins produced by either normal or tumor cells into the tumor microenvironment affect the malignant behavior of the tumor cells (11).
In order to clarify the molecular mechanisms involved in the development and progression of breast cancer and to discover new markers with diagnostic and therapeutic values for the disease, in recent years, many attempts have been made to examine the differential expression of proteins by breast cancer cells and normal cells, using different proteomic approaches including; sodium dodecyl sulfate (SDS)-polyacrylamide or two dimensional polyacrylamide gel electrophoresis (2D-PAGE), surface enhanced laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS/MS), high performance liquid chromatography (HPLC), etc. and using various samples (12, 13).
2. Objectives
Although there is very limited evidence about the differences between proteome of breast cancer tissue and the adjacent normal tissue (11), to the best of our knowledge, this subject has not gone under investigation in luminal A (ER+, PR+, and HER2-) breast carcinoma. Thus, the present study was conducted to characterize and compare the proteome of these different nests in the Iranian women with 3rd stage of luminal A breast ductal carcinoma.
3. Methods
3.1. Patients and Specimens
Study population consisted of 5 women with 3rd stage of breast cancer undergoing curative breast mastectomy in the Hospital of Zanjan University of Medical Sciences from February, 2015 to May, 2015. Patients with history of ovarian cancer, and those who had received medication, radiotherapy, etc., were not included. During the excision surgery, 50 mg of fresh breast cancer tissue as well as 50 mg of the adjacent normal breast tissue were obtained from each patient. Discrimination between breast cancer tissues and the adjacent tissues was made by pathological examinations. In order to remove blood remnants and prevent their interference with the tissue protein content, the tissues were washed with phosphate-buffered saline (PBS) under the sterile condition, and were immediately saved at -70°C.
3.2. Tissue Protein Extraction
Prior to protein extraction, the tissues were separately crushed and homogenized with liquid nitrogen. Then, 400 µL of lysis buffer solution [containing 6 M urea, 2 M thiourea, 50 mM Tris, 50 mM DTT, and 2% (w/v) CHAPS] with 10 mM PMSF (with a ratio of 1 to 20) and P8340 (with a ratio of 1 to 20) was added (per 150 mg of sample), incubated at room temperature for one hour, followed by centrifugation at 14000 g at 4°C for 15 minutes, the supernatant was collected, and the protein concentration of each supernatant was measured by Bradford protein assay, as previously described (14). Finally, a cancer protein pool was prepared by using 200 mL of protein solution extracted from each breast cancer sample, and so was done for preparing a normal tissue protein pool to minimize the error rate.
3.3. 2D-PAGE Electrophoresis
The 2D-PAGE electrophoresis allowed the separation and analysis of protein content of normal and tumor tissue protein pools as follows: first, 100 ml of each pool was dissolved in 150 mL of an aqueous buffer (containing 8 M urea, 50 mM DTT, 0.0002 Bromophenol blue, 0.5% Biolyt 3-10 and 4% CHAPS), in order to increase the solubility of the extracted proteins, centrifuged at 14000 g at 4°C for 10 minutes, and the non-linear immobilized pH gradient (IPG) strips (pH 3-10, 11 cm; Bio-Rad, USA) were placed into it (in triplicates). After one hour, the strips were covered with mineral oil and incubated at room temperature for 16 hours. The first-dimension isoelectric focusing (IEF) was then carried out using Ettan IPGIV (GE Health care UK Limited, Amersham Place, UK) under the following conditions: linear1, 500 V for 60 minutes; gradient1, 1000 V for 60 minutes; gradient2, 6000 V for 150 minutes; and linear2, 6000 V for 30 minutes. Before performing the second-dimension (SDS-PAGE) analysis, the IPG strips were first equilibrated (recharged) in two equilibration buffers to allow the proteins to move onto the SDS-PAGE gel. Briefly, the strips were equilibrated in the first buffer containing; 6 M urea, 50 mM Tris- HCL, 2% (w/v) SDS, 30% (w/v) Glycerol, and 1% (w/v) Dithiothreitol (DDT) for 20 minutes, and then in the second buffer containing; 6 M urea, 50 mM Tris- HCL, 2% (w/v) SDS, 30% (w/v) Glycerol, and 4% (w/v) Idoacetamide for another 20 minutes. For SDS-PAGE analysis, the equilibrated strips were placed on the 12% SDS polyacrylamide gels and fixed with in situ using 0.5% low-melt agarose (Merck, USA). The SDS-PAGE was then carried out under the following conditions: 15 mA for 60 minutes and 25 mA until the Bromophenol blue color reached the end of the gel (6 - 7 hours).
3.4. Gel Staining
After the 2D-PAGE electrophoresis, the gels were placed into a Coomassie brilliant blue G250 solution (Merck, USA) for 3 - 4 hours, and washed twice with bleaching solution for 20 minutes, followed by one-night incubation in the bleaching solution to increase transparency of the gels.
3.5. Gel Analysis and Protein Identification
The stained gels were then scanned by image scanner IV (GE Health care), and were analyzed using ImageMaster 2D Platinum software version 6.0 (GE Health care). The differences in protein contents between expressed proteins in cancer breast tissues and the adjacent normal tissues were preliminarily examined by the software. Then, the spots of interest were identified by MALDI-TOF MS/MS at the University of York (England).
3.6. Ethics
Adherence to the ethical principles of studies on human samples was approved by the ethics committee based in Zanjan University of Medical Sciences. Also, all patients signed a written informed consent regarding the use of their samples and clinical data. No financial expense was imposed on the patients.
3.7. Statistical Analysis
To examine the differences in protein content of the groups, an independent t test was performed using ImageMaster 2D Platinum software. Also, the GraphPad Prism 5 software for Windows (San Diego California, USA) was used for developing a graph. Within the confidence interval of 95%, the P values < 0.05 and < 0.07 were considered significant and at the borderline of significance, respectively.
4. Results
A total of 5 patients were included, all of which were pathologically confirmed to be at the 3rd stage of luminal A (ER+, PR+, and HER2-) ductal carcinoma. The mean age of patients was 47.0 ± 10.9 years. Out of 5, 4 patients were premenopausal and one patient was postmenopausal.
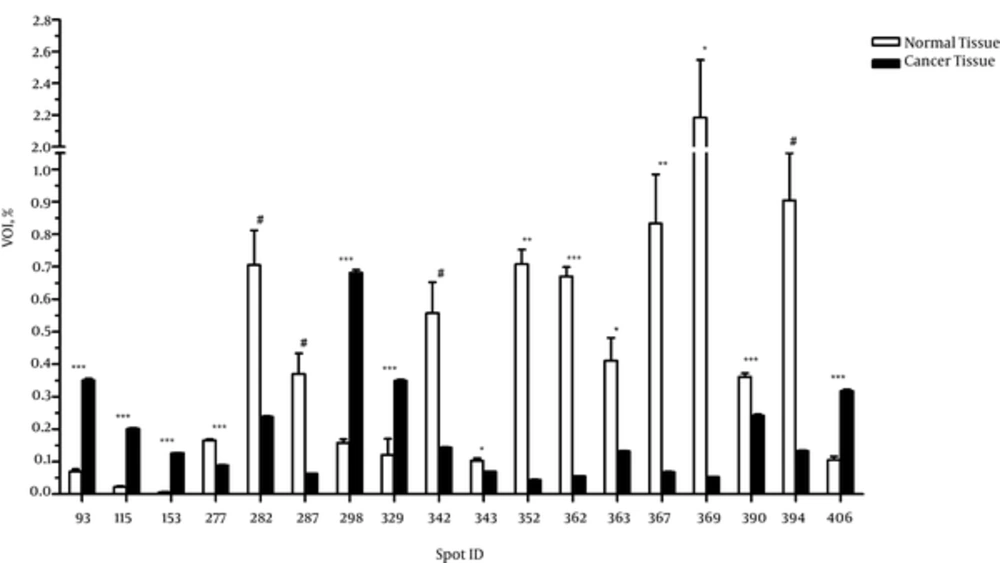
Analysis of 2D gel images of from both breast cancer and the adjacent normal tissues revealed about 1300 spots in the breast cancer tissues and 751 spots in the adjacent normal tissues, among which 46 spots were matched between the cancer and normal tissues. In addition, 1254 out of 1300 spots were found exclusively in the cancer tissue, considered as novel spots, and 705 out of 751 spots were only detected in the adjacent normal tissues, considered as deleted spots. According to the results of the independent t test, 14 out of 46 matched spots showed significantly different expression between the cancer and adjacent normal tissues (P < 0.05), and 4 spots exhibited differences at borderline significance (Figure 1).
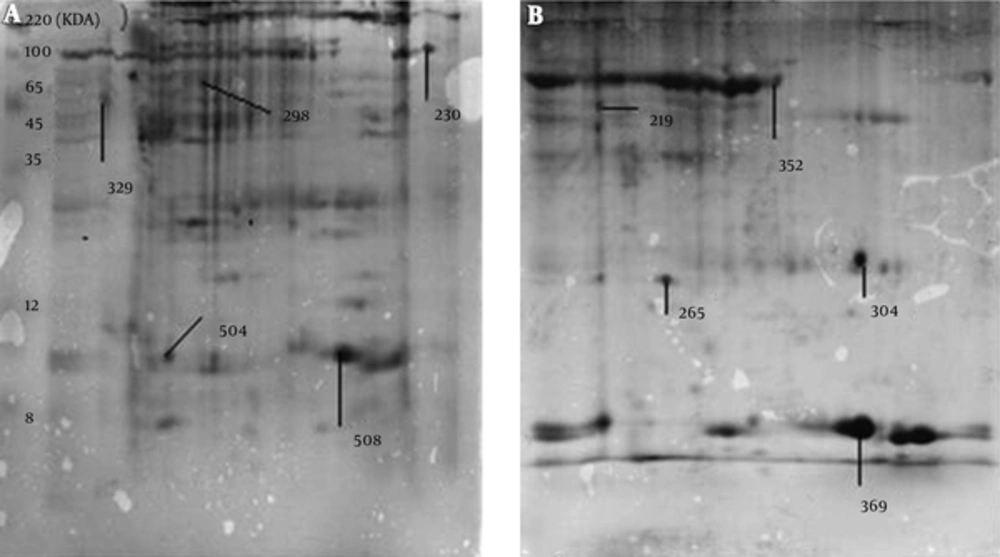
The MALDI-TOF MS/MS analysis were used for protein identification of 3 novel spots and 2 deleted spots as well as of 4 matched spots with different expressions between the cancer and adjacent normal tissues (Figure 2). According to the results of MALDI-TOF MS/MS analysis, calreticulin, galectin-1, and constant region of Ig gamma-1 chain were identified in the spots belonging to breast cancer tissue, and alpha-1-antitrypsin and hemoglobin subunit beta were identified in the spots belonging to the adjacent normal tissue (Table 1). Additionally, one of the spots with approximately 4 fold increased expression in the breast cancer tissue compared to the adjacent normal tissue (P < 0.001) was identified as a single peptide matching to cytoskeletal keratin type I and type II. The other spot that showed approximately 2 fold increase in the breast cancer tissue (P < 0.001) was identified as C region of Ig kappa chain (Table 2). Moreover, one of the spots that had approximately 16 fold reduction in the breast cancer tissue compared to the adjacent normal tissue (P = 0.007) was attributed to serum albumin and a single peptide matching to Collagen alpha-3(VI) chain. The other spot with approximately 41 fold decrease in the breast cancer tissue (P = 0.04) was attributed to tropomyosin alpha-4 chain and a single peptide matching to putative tropomyosin alpha-3 chain-like protein (Table 2).
| Match ID | Protein | Tissue |
|---|---|---|
| 219 | Alpha-1-antitrypsin | Normal |
| 304 | Hemoglobin subunit beta | Normal |
| 230 | Calreticulin | Cancer |
| 504 | Ig gamma-1 chain C region | Cancer |
| 508 | Galectin-1 | Cancer |
Proteins Detected Exclusively in Cancer Tissue or the Adjacent normal Tissue by MALDI-TOF MS/MS Analysis
| Spot ID | Proteins Identified | Folds (C/N) | t | P Value |
|---|---|---|---|---|
| 298 | Single peptide matching to cytoskeletal keratin type I and type II | 4.32 ↑ | 31.20 | 0.000 |
| 329 | Ig kappa chain C region | 1.98 ↑ | 22.00 | 0.000 |
| 352 | Serum albumin and a single peptide matching to Collagen alpha-3(VI) chain | 15.97 ↓ | -12.13 | 0.007 |
| 369 | Tropomyosin alpha-4 chain, a single peptide matching to putative Tropomyosin alpha-3 chain-like protein | 41.37 ↓ | -4.82 | 0.040 |
Differences in Intensity of Matched Spots Between Cancer Tissue and the Adjacent Normal Tissue, Values are Presented as Mean ± SD.
5. Discussion
In the present study, three proteins were exclusively detected in breast cancer tissue including calreticulin, galectin-1, and constant region of Ig gamma-1 chain. In addition, the expression of cytoskeletal keratin types I and II, and C region of Ig kappa chain were fortified in the cancer tissue compared to the adjacent normal tissue. On the other hand, the alpha-1-antitrypsin and hemoglobin subunit beta were exclusively observed in the normal adjacent tissue. Furthermore, abundance of serum albumin, collagen alpha-3 (VI) chain as well as tropomyosin alpha-4 and alpha-3 chains in the cancer tissue was significantly lower than that of the adjacent normal tissue.
Galectin-1 is a well-known lectin expressed in variety of malignant tumors (15). Although galectin-1 role was primarily described in tumor cell adhesion (16), it later turned out to be involved in apoptosis induction of activated T cells by tumor cells (17). Also, Butler et al. (18) have suggested galectin-1 as a substrate for matrix metalloproteinase 14 (MMP14), which is expressed by different types of breast carcinomas. In addition, galectin-1 has been reported to regulate tumor angiogenesis (19). What’s more, Lahm et al. (20) demonstrated that various types of breast cancer cell lines express galectin-1. Additionally, it is shown that galectin-1 expression in tumor microenvironment is associated with increased apoptosis of T cell (21), as well as with tumor invasiveness and progression in breast cancer (22). Moreover, Imai et al. (23) observed that the expression of galectin-1 is increased in human metastatic breast cancer cells compared to non-invasive cancer cells and normal cells. Also, Daroqui et al. (24) demonstrated that galectin-1 is upregulated by TGFβ in metastatic breast adenocarcinoma. Furthermore, Mackay et al. (25) showed that overexpression of (HER2/neu) in immortalized human mammary luminal epithelial cells is associated with enhancement in the expression of 35 genes, including galectin-1. Thus, the result of the present study indicating the expression of galecion-1 in the breast cancer tissue is consistent with preceding evidence and supports the concept regarding the role of galecion-1 in breast tumorogenesis and tumor progression.
The function of immunoglobulins (Igs) in context of cancer is not well elucidated. However, previous evidence suggests that presence of Igs in the serum is associated with anti-tumor immune suppression (26) and tumor progression (27). In addition, overexpression of constant regions of Ig gamma-2 and Ig mu chains in breast cancer has been recently documented by Cho et al. (28). More recently, it has been reported that carcinomas and other cancer cells produce IgG (29) that contributes to cell proliferation, progression (30) and metastasis (31). In the present study, the expression of constant region of Ig gamma-1 and Ig kappa chains increased in the cancer tissue compared to the adjacent normal tissue, which is in agreement with the findings of Cho et al. (28). Howbeit, Deng et al. (10), in contrast to the present study, observed that the expression of Ig heavy and light chains was down-regulated in the breast cancer tissue compared to the adjacent normal tissue. Thus, clarification of the Igs’ role in the pathogenesis of breast cancer requires intensive work.
Calreticulin is multifunctional Ca2+-binding chaperone (32) known to crucially be involved in regulation of gene transcription (33), accurate protein folding (34), and also apoptosis induction in breast cancer cells (35). The finding of the present study regarding expression of calreticulin in cancer nest but not in the normal adjacent tissue contradicts previous reports (36, 37). Peng et al. (36), for instances, reported that expression of calreticulin was decreased in the colon cancer tissue compared to the adjacent normal epithelium, which was correlated with diminished infiltration of CD45RO+ cells to the cancer nest. However, interestingly, Eric et al. (38) have shown that simultaneous presence of stromal IgG immunoreactivity and overexpression of calreticulin is associated with increased metastasis in breast cancer. Thus, with respect to expression of calreticulin and overexpression of Igs in the breast cancer nest, the present study is in agreement with Eric et al. (38) and suggests that an aberrant expression of calreticulin might be implicated in breast cancer pathogenesis.
The cytoskeletal keratins play structural, protective, signaling and regulatory functions (39). Several studies have accumulated evidence to suggest that keratins are involved in cancer cell progression, metastasis, and response to treatments (40). As such, their clinical utility as reliable diagnostic tumor markers in a variety of epithelial cell carcinomas (41), including breast cancer (42) has been drawing attentions of many scientists and clinicians. The result of the present study showing increased expression of cytoskeletal keratin types I and II in the breast cancer tissue is in line with previous studies (43). Somiari et al. (43), for instance, observed that cytokeratin-18, a member of the type I keratin family, was overexpressed in the breast cancer microenvironment, correlated with poor prognosis of the disease. More recently, Cheung et al. (44) found that keratin 14, another member of the type I keratin family, is a key regulator of polyclonal breast cancer metastasis so that keratin 14-expressing tumor cells are the main origin of metastatic cells.
The type VI collagen has been known as an anti-apoptotic and cell-proliferation stimulating factor (45). For example, it has been proposed that the adipocyte-derived collagen VI promotes breast tumor growth and progression (46). However, in the present study, the collagen VI revealed to be derogated in the breast cancer nest compared to the adjacent normal tissue. Although the underlying mechanism remains uncertain, according to Sato et al. (47), the increased expression of MMPs that degrade the collagen IV in advanced metastatic cancers, might be responsible for diminution of this protein, observed in the present study.
Another controversial and inconclusive observation of the present study was that unlike the previous reports, the expression of tropomyosin alpha 3 and alpha 4 chains, and alpha 1-antitrypsin was waned in the breast cancer tissue. In other words, plethora of literature suggests mounted expression of these oncogenes in lymphomas (48) and carcinomas (49, 50), as well as in breast cancer (51). Gromov et al. (52) in a proteomic study, for instances, found that tropomyosin alpha 3 and alpha 4 chains were up-regulated in the breast tumor microenvironment. In addition, Lopez-Arias et al. (51) reported an augmented alpha 1-antitrypsin as a novel tumor-associated biomarker for early-stage breast carcinomas. Interestingly, consistent with our study, Deng et al. (10) observed that the expression of alpha 1-antitrypsin was down-regulated in the breast cancer tissue compared to the adjacent normal tissue. Although it is uncertain yet, the present study in accordance with that of Deng et al. (10) suggests that the overexpression of alpha 1-antitrypsin in the cancer nest, independent of its serum level, might be implicated in the pathogenesis of breast cancer.
In the present study, the expression of albumin was reduced in the breast cancer net, and the hemoglobin subunit beta was not detected in the breast cancer tissue, while it was seen in the adjacent normal tissue. These observations are consistent with previously provided evidence (53). By way of example, reported that the serum level of albumin can be implemented as an independent predictor for survival of breast cancer patients (54). Additionally, down-regulated expression of serum albumin in the breast cancer tissue has been previously reported by Deng et al. (10). Since the albumin and hemoglobin are important proteins of blood and play very crucial roles in blood transportation of materials, oxygen and Carbone dioxide as well as in maintaining the colloid osmotic blood pressure, these finding suggest that blood support in breast cancer tissue is less than the adjacent normal tissue. Also, based on previous evidence (55), the reductions in concentration of albumin and hemoglobin in the cancer tissue might be due to catabolism of these proteins by the tumor itself.
The present study adds considerable data to the literature in many aspects regarding the molecules possibly involved in the pathogenesis of breast cancer, as discussed above. However, as weakness of the present study, proteins of all matched spots between the cancer and normal tissues were not identified by MALDI-TOF MS/MS analysis, due to technical and financial limitations. Moreover, the use of non-linear pH IPG strips did not allow further comparison of majority of the novel and deleted spots with their online counterparts. Therefore, these limitations should be taken into account in future investigations.
In conclusion, it seems that overexpression of galectin-1, the types I and II keratin, as well as down-regulation of collagen VI and alpha 1-antitrypsin in the cancer nest might be implicated in the pathogenesis and progression of advanced breast carcinoma. In addition, despite the difference in the expression of calreticulin, immunoglobulin chains, alpha 3 and alpha 4 tropomyosin in the breast cancer tissue compared to adjacent normal tissues, these changes do not seem to harbor diagnostic value for the disease, as they contradict previous evidence. However, much more studies are suggested to confirm our results and also to clarify the underlying mechanism.