1. Background
Tongue Squamous Cell Carcinoma (TSCS) is one of the important aggressive head and neck cancers whose management and treatment is challenging (1). This tumor is frequent in people with chronic smoking, drinking, and betel squid chewing habits. The clinical symptoms are mostly apparent at the late stage of the disease when the chance of mortality is high; in addition to this, there is a high possibility of recurrence of malignancy after the treatment interventions (2, 3). Despite improvement of disease research and management, these measures are still inadequate due to the disease complexity (3). So, a need for reaching a more sophisticated method of disease management is needed. In this light, biomarker investigation is helpful to better understand the nature of the disease and consequently to achieve novel prognosis and treatment approaches. Transcriptomic, microRNA profiling and proteomics are the most recent high-throughput evaluations of TSCS (2, 4, 5). Specifically, proteins as the fundamental acting parts of living organisms have a definite place in molecular research. Many abnormal phenotypes and disease onset and developments are linked to the proteins’ behaviors (6). Furthermore, interpretation of interaction of these fundamental elements can provide additional knowledge about the whole system malfunctions. In the other words, organism functions are facilitated through complex molecular interactions (7). In a protein-protein interaction network, there are complex interactions between proteins where any alteration in this level can promote an abnormal phenotype such as a disease condition (8). In fact, any small modifications in these agents may affect the molecular interaction arrangements and biological processes that are known as key for normal functions (9). In this regard, network analysis is a useful tool to introduce the crucial proteins as essential elements of the network among numerous proteins. This is possible via topological evaluation of the each protein (10). Degree, betweenness, and closeness are used frequently as the important centrality parameters (11). Degree is the number of the edges that are ended to a node (protein) and the node with a high value of degree is called hub (12). Betweenness value for node T in a network including N nodes is calculated as ratio of G phrase divided by the number of the possible paired nodes excluding the T node (13). Where G is sum of the ratio of the shortest paths between a paired node, where a node acts as bridge in those divided by the all shortest paths between these two nodes. The node characterized by high value of betweenness is known as bottleneck node. Closeness, the other centrality parameter of a node T, is equal to inverse of average of shortest paths between the node T and the other nodes (14). These highlighted parameters via topological analysis may play a considerable role in disease mechanisms (15). Therefore, in this study by the application of protein-protein interaction network, an attempt is made to provide a preliminary knowledge of interaction pattern and to introduce crucial interacting biomarker panel of tongue squamous cell carcinoma.
2. Methods
The protein-protein interaction network was constructed and analyzed by Cytoscape 3.5.1 and its applications. In a way that the network query was by STRING DB V 10.5 (http://string-db.org/), Plug-in. Three different sources are applicable through string DB including protein query, PubMed query, and disease query (16). The latter one was used for the network construction and a confidence score of interaction was set to 0.4. The topological understanding of the network was by the application of Network Analyzer, which is a well-integrated application in Cytoscape (17). Three critical centrality parameters were focused for the network evaluation including degree (K), betweenness centrality, and closeness centrality. Nodes with highest values of degree and betweenness centrality are considered as hub and bottleneck elements, respectively (10). Following the topological analysis, the important nodes were targeted for enrichment examination and action type identification via ClueGo + CluePedia Plug-ins. STRING Action File available in the CluePedia Panel was the source for action query. The action type analyses were activation and inhibition which are assigned with different colors. The statistical method for scoring the actions was kappa statistics which is customizable from 0 to 1 and it can be shown as thick and thin lines. Here, it is set to 0.5 (medium) cut off for any kinds of actions. Gene ontology analysis of the essential elements was based on biological process identification. Clusters of terms are connected with the assigned kappa scores in biological processes analysis. The higher the kappa score, the higher the possibility of terms’ linkage. The assigned statistical criteria are as below:
Kappa score = 0.4, Corrected P value < 0.05
Number of genes per term = 3, Percentage for the queried terms = 4
Grouping level: Min = 2, Max = 8
P value correction method: Bonferroni step down. Enrichment/depletion test: two-sided (enrichment/depletion) test based on hypergeometric distribution (18, 19).
3. Results
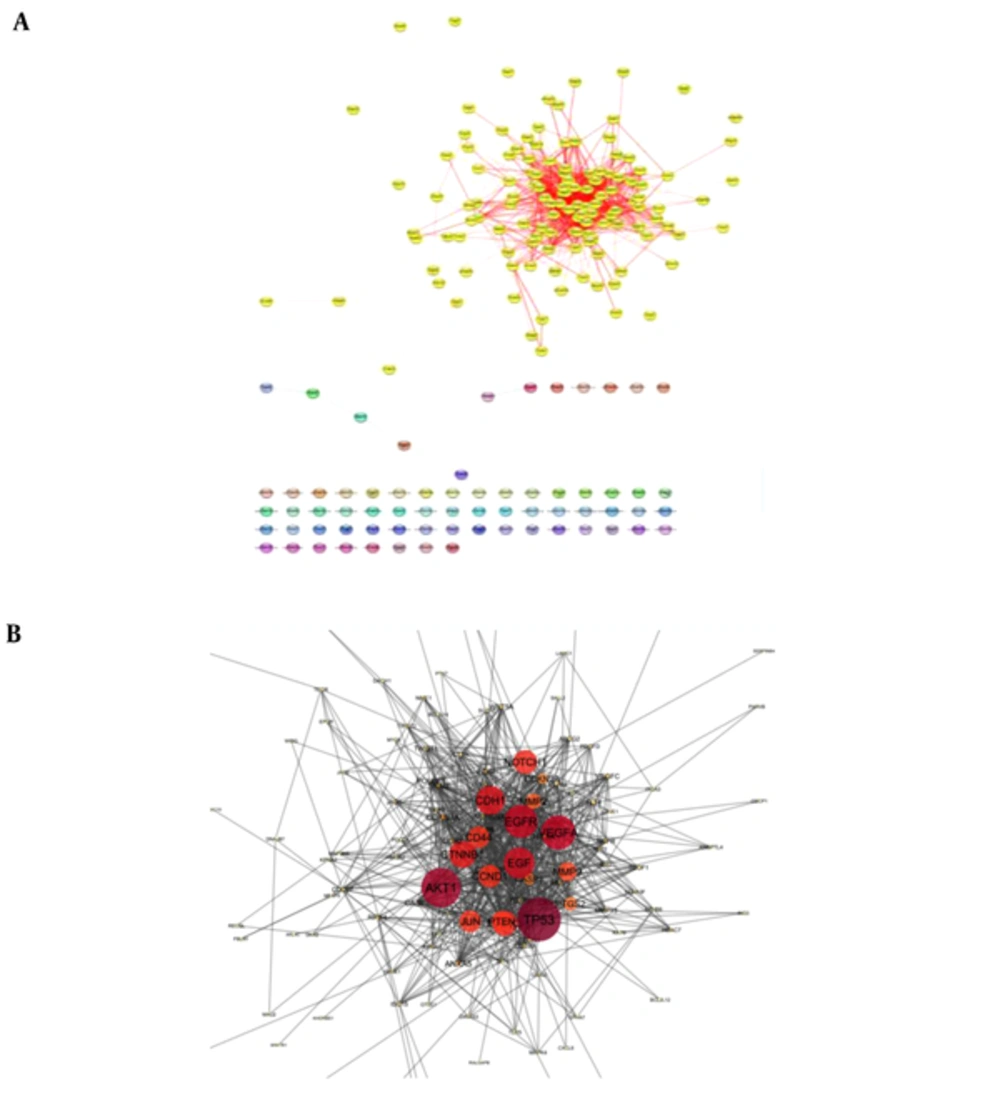
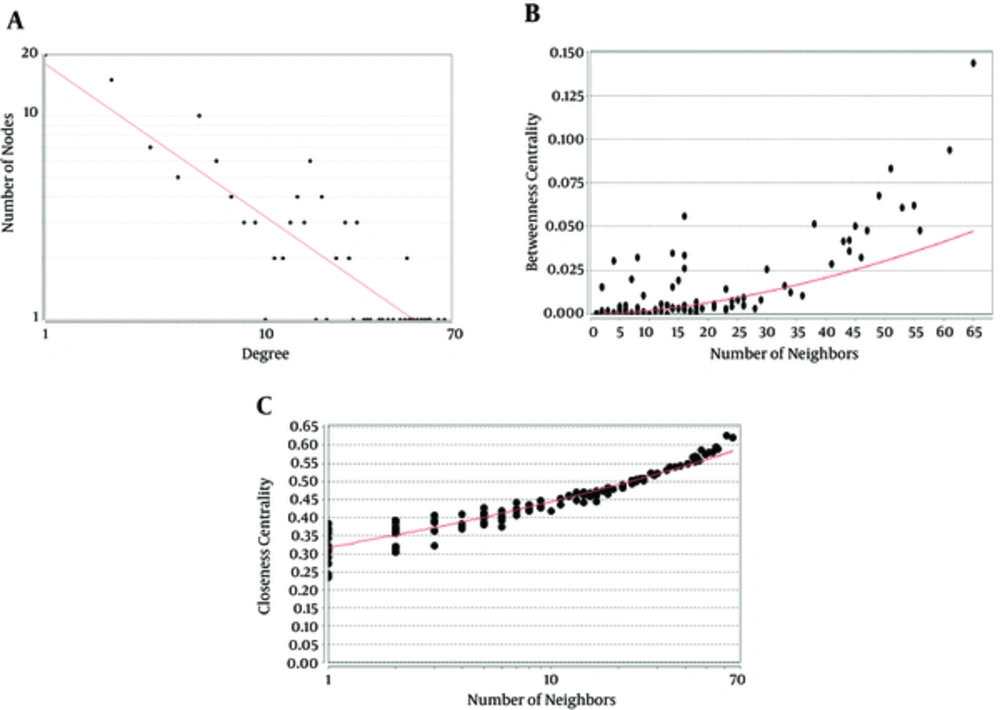
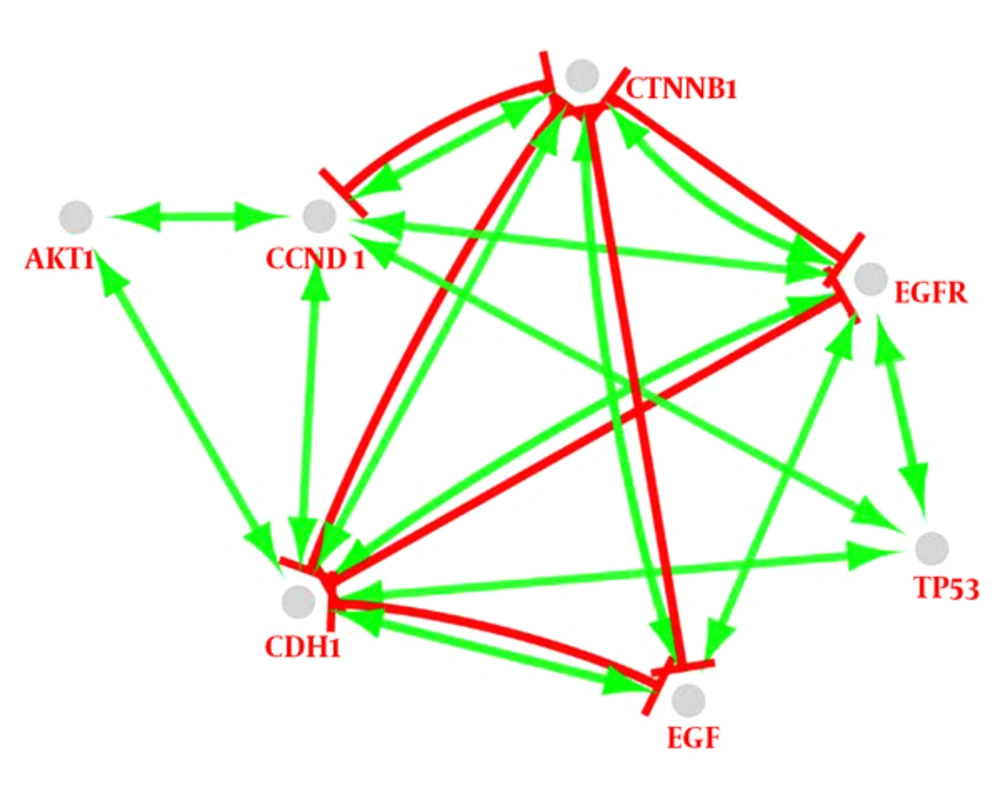
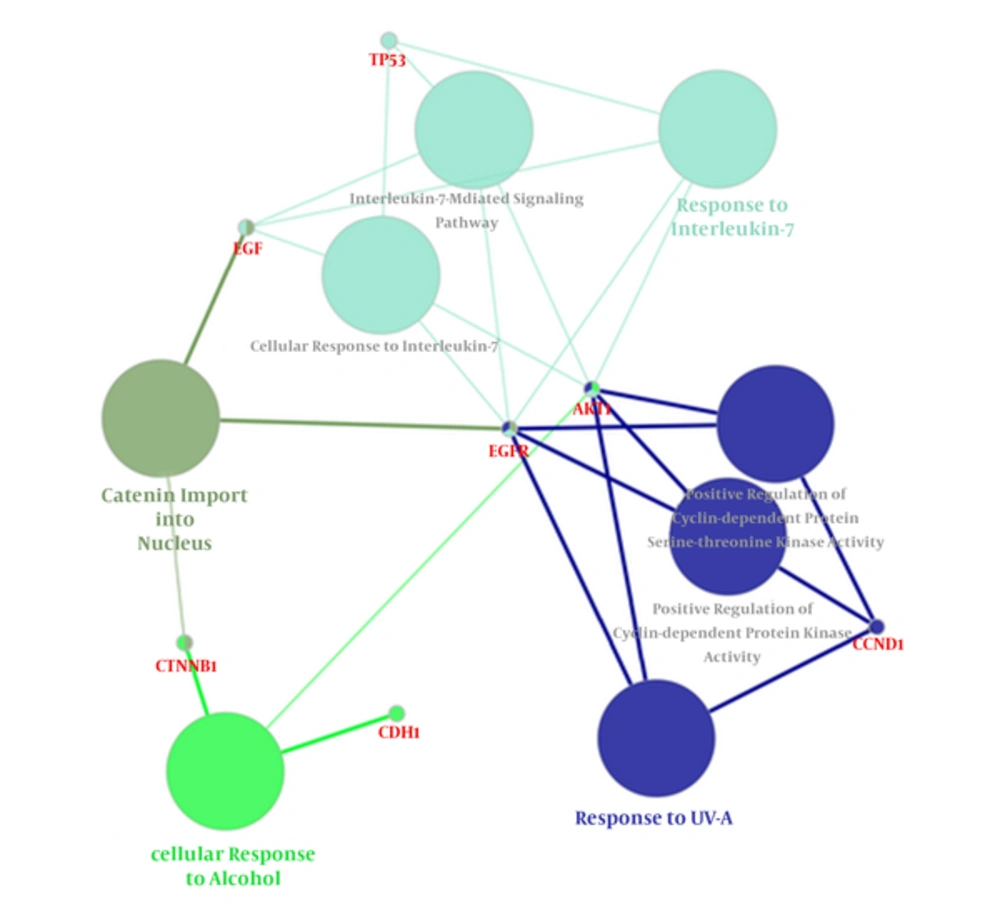
Following the network query for 200 related proteins in tongue squamous cell carcinoma via string DB, Cytoscape, a subnetwork of connected nodes was extracted (Figure 1). Analyzing the distribution of important parameters of the network including degree, betweenness, and closeness centrality is presented in Figure 2. The finding corresponds to the scale free network. Cut offs for degree, betweenness centrality and closeness centrality were determined based on average + 2SD (20). In the mentioned article, the applied method was only suggested for degree value, but here we have adjusted it for other centrality values as well. By this method the suitable number of nodes were selected. So the critical nodes were selected by degree, betweenness, and closeness values above 45, 0.05, and 0.56, respectively (Table 1). Interaction type analysis can provide further information about the central proteins’ behavior in the constructed network (Figure 3). Enrichment analysis based on biological process of the crucial proteins, as it is shown in the Figure 4, can provide information related to disturbed processes and their roles in the disease pathogenicity. The reason for choosing only critical proteins for gene ontology analysis was to determine the more essential biological processes.
| Row | Name | Description | D(K) | BC | CC | DS |
|---|---|---|---|---|---|---|
| 1 | TP53 | Tumor protein p53 | 65 | 0.14 | 0.62 | 1.30 |
| 2 | AKT1 | V-akt murine thymoma viral oncogene homolog 1 | 61 | 0.09 | 0.62 | 0.94 |
| 3 | EGFR | Epidermal growth factor receptor | 55 | 0.06 | 0.60 | 0.80 |
| 4 | EGF | Epidermal growth factor | 53 | 0.06 | 0.58 | 0.70 |
| 5 | CDH1 | Cadherin 1, type 1, E-cadherin (epithelial) | 51 | 0.08 | 0.57 | 1.66 |
| 6 | CTNNB1 | Catenin (cadherin-associated protein), beta 1, 88kDa | 49 | 0.07 | 0.57 | 1.34 |
| 7 | CCND1 | Cyclin D1 | 45 | 0.05 | 0.56 | 1.02 |
aCut offs for degree, betweenness centrality and closeness centrality are 45, 0.05, and 0.56, respectively. The disease score is indicated as DS.
A, A network of tongue squamous cell carcinoma including 132 interacting and 68 isolated nodes is presented. The connected elements are highlighted in yellow while the individuals are in different ranges of colors. B, the main connected component including 132 nod and 923 edges is illustrated.
A Scale Free Network: Nodes Are Shown as Black Circles and the Red Line Indicates the Power Law. A, Degree distribution, Correlation: 0.94 and R-squared: 0.76; B, Betweenness Centrality distribution, Correlation: 0.85 and R-squared: 0.45 and C, Closeness distribution, Correlation: 0.95 and R-squared: 0.86.
Clusters of Biological Processes Related to Central Elements of the PPI Network. Different groups are coded with specific colors. Response to UV-A, Response to interlukin-7, cellular response to alcohol, and catenin import into nucleus process are the identified significant processes with their corresponding central proteins of the network. Kappa score = 0.4, Corrected P value < 0.05.
4. Discussion
Network medicine is a relatively new discipline that applies different algorithms to provide a better understanding of molecular concept of different diseases including cancer (9). Cytoscape as one of the most popular software for network construction (see Figure 1) and characterization is selected for the present study. With regard to the complex structure of interacting map, more information can be reachable through topological assessments. For this reason, the connecting nodes were examined for the further study by the relevant plug-ins. Node distribution analysis (Figure 2) indicates a scale free mode of the network. In scale free networks, the nodes play different roles in the network and can be ranked as crucial and ordinary nodes in the studied network (21). The trend in degree, betweenness, and closeness centralities distribution implies on noteworthy distance of limited numbers of proteins from the other ones. Considering calculated cut offs for assessing centrality parameters based on designed method (22), seven proteins including TP53, AKT1, EGFR, EGF, CDH1, CTNNB1, and CCND1 were purposed as indicated in Table 1. These proteins were reviewed by literature survey all of which were confirmed to have significant associations in TSCS by many studies (23-28). As it will be discussed, these proteins have been known to have noteworthy participation in many malignancies. Therefore, a panel of them may be more promising in tongue cancer screening. The important role of TP53 in prognosis of TSCC is discussed in details (29). However this protein is related to almost all of the cancer types (30). Based on previous investigations, dysregulation of AKT1 is linked to TSCC. Its role in lymph node metastasis is highlighted. Correlation between AKT1 and reduced overall survival of the cancer patient is also reported (31, 32). There is compelling evidence about involvement of EGF and EGFR in numerous cancers such as colorectal, breast, osteosarcoma, and gastric cancers (33-35). Increment of gene expression of EGFR in oral tongue squamous cell carcinoma was reported by M. Ryott et al. (2009) (25). The linkage of CDH1 is studied in the several cancers including TSCC (36-38). The prominent roles of CTNNB1 in nasopharyngeal carcinoma, tongue cancer cell growth, migration and invasion, initiation of some colon carcinomas and melanomas and prostate cancer are reported in some studies (28, 39-41). There is a correlation between leukemia, primary head and neck squamous cell carcinoma including tongue cancer, and lung cancer with CCND1 expression (42-45).
As it is shown in the Figure 3, there is a complex inhibitor or activator interaction between the seven key genes. TP53 and AKT1 are activated merely by the other genes directly or indirectly but the other genes are activated and also inhibited. Since TP53 activation can induce apoptosis or cell-cycle arrest (46), it seems that the elements of biomarker panel are organized against cancer progress and development. Additionally, these proteins are in condensing interactions as indicated in Figure 3. CDH1 and CTNNB1 have most actions in this network. In spite of the key role of CTNNB1 in the interaction with other genes, it does not have a direct relationship with TP53 and AKT1. However, an investigation indicates that TP53 mutation frequently is accompanied by CTNNB1 mutation (47).
In Figure 4, gene ontology properties of the recognized central proteins based on biological process are illustrated. Response to UV-A, response to interlukin-7, cellular response to alcohol, and catenin import into nucleus were assigned to our top central proteins. That is, these processes may undergo vast modifications as the central proteins dysregulate. The connection of these processes with tongue cancer can be explained in a way that, among these processes, only Response to UV-A has not yet been addressed by any previous studies. Cellular response to alcohol supports the fact that alcohol has some links to this type of cancer as was previously reported about the role of alcohol in oral cancers (48). Association of interleukins with tongue squamous cell carcinoma has been previously reported (49, 50). As mentioned earlier, Catinin contributed to tongue cancer development and invasion; therefore, catenin import into nucleus relation can be justified (28).
In conclusion, the introduced potential biomarker panel and their associated biological processes in this study can be important for more evaluation and consequently for the tongue squamous cell carcinoma treatment goals.