1. Context
Breast cancer is the most common cancer in women worldwide (1). Hormone receptor positive breast cancer constitutes the majority of these cancers, with positive nuclear staining for estrogen receptor (ER) in 75% and progesterone receptor (PR) in 55% of tumors (2). The role of estrogen in breast tumorigenesis and cancer progression is well recognized. This link was first described in the 19th century when Beatson used oophorectomy to treat women with advanced breast cancer (3). Overall, ER positive tumors are more common than ER negative tumors in all age groups. However, the proportion of hormone positivity increases with age, such that breast tumors in postmenopausal women are overwhelmingly ER positive (4, 5). On the molecular level, ER positive breast tumors mainly constitute the luminal A and luminal B subtypes. Luminal A tumors mostly co-express ER and PR and are associated with a better prognosis compared to luminal B tumors (6).
Endocrine strategies for treatment of ER positive breast cancers include targeting estrogen receptor by the anti-estrogens tamoxifen and fulvestrant, or suppressing the amount of available ligand (estrogen) for the receptor, either with aromatase inhibitors (AIs) in postmenopausal women, or gonadal suppression in premenopausal women (luteinizing hormone-releasing hormone agonists or ovariectomy). Hormone therapy has significantly improved clinical outcomes for early- and late-stage hormone receptor positive breast cancer (7). Although most patients with early stage breast cancer are treated with curative intent, approximately 20% - 30% of patients eventually experience a recurrence (8). Endocrine resistance in ER positive breast cancer can be either due to de novo resistance (i.e. patients do not respond to first line endocrine treatment) or acquired resistance (ie. patients will eventually experience a relapse despite an initial response). During the last two decades, there have been tremendous efforts to understand the biological mechanisms of hormone therapy resistance, with the ultimate goal of implementing new therapeutic strategies to improve the current treatments for ER positive breast cancer (7, 9-12). Several mechanisms of hormone therapy resistance have been proposed, including genetic alterations that lead to altered ER expression or ERs with changed protein sequence. It has been suspected for many years that acquired ER mutations which happen after initiation of hormone therapy may play a role in treatment failure and breast cancer progression. By altering the conformation of the receptor and augmenting its interaction with coactivators, point mutations in ESR1, the gene encoding estrogen receptor α, can promote ER activity in the absence of hormone which ultimately assists tumor cells to evade hormone therapy (13). In this review, we will address the current knowledge about estrogen receptor, its genomic alterations during breast cancer evolution and their proposed role in hormone therapy resistance.
1.1. Estrogen Receptor Structure and Function
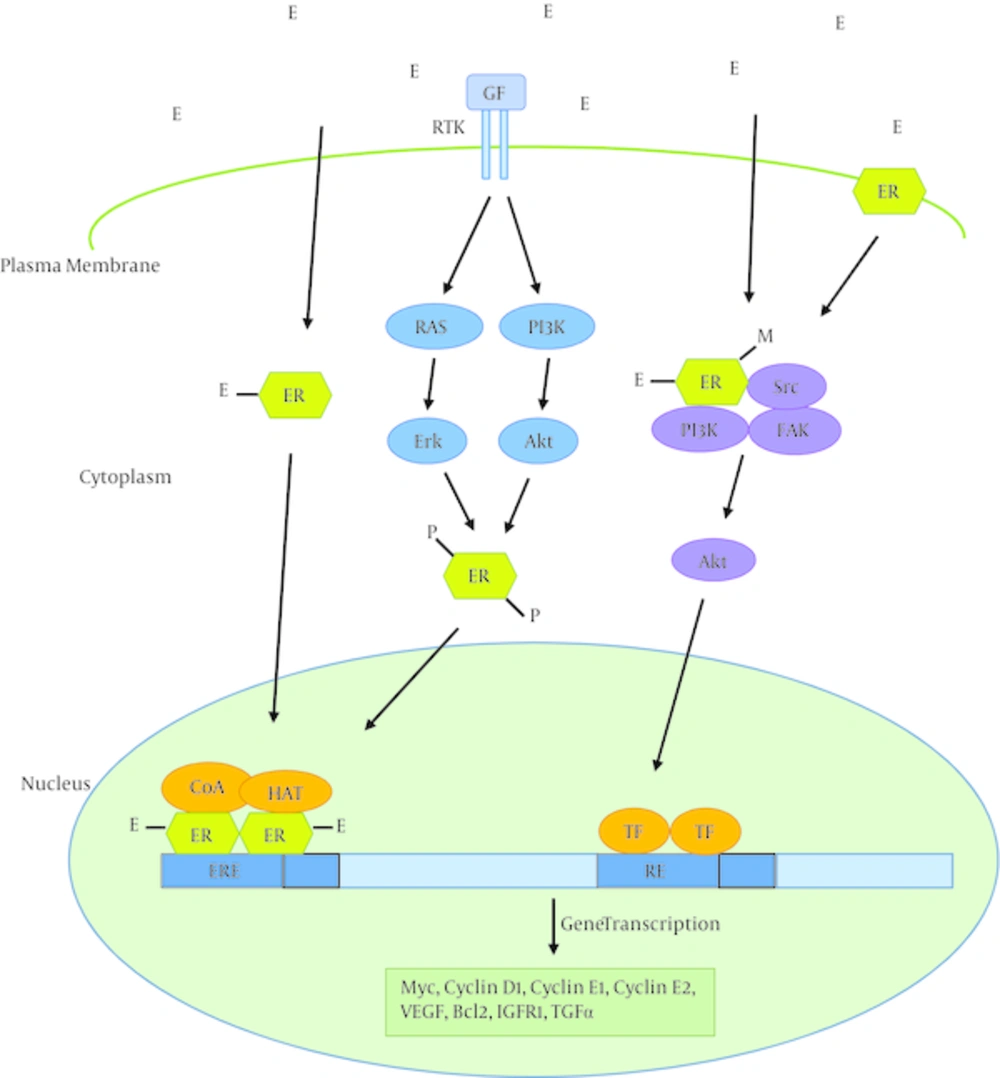
Estrogens perform their function through interaction with estrogen receptors. Estrogen receptors are members of the superfamily of nuclear hormone receptors including those of other steroid hormones, thyroid hormone, retinoid acid and vitamin D. There are two estrogen receptors: ERα (encoded by ESR1) and ERβ (encoded by ESR2). These two receptors have similar, but not identical, structures. ERα mediates many of the effects of estrogens on normal and cancerous breast tissue via ligand activated regulation of gene expression (genomic actions) and by functioning as a member of membrane and cytoplasmic signaling pathways (non-genomic actions) (Figure 1) (9).
Pathways of estrogen regulation of gene expression have been shown. In the classical estrogen signaling, estrogen (E) binds estrogen receptors (ERs), induces dimerization of the protein, and activates gene expression through binding to estrogen response elements (EREs) in the promoter of target genes, in complex with co-activators (CoAs) and histone acetyl transferases (HATs). Activation of signaling events downstream of receptor tyrosine kinases (RTKs) can also lead to ER phosphorylation (P) through Erk or Akt serine/threonine kinases and subsequent ligand-independent activation of ER. Estrogen signaling can also be mediated through non-genomic mechanisms by ER in the cytoplasm or membrane. Ligand binding leads to the formation of functional protein complexes that involve other signaling pathways, resulting in transcription factor (TF) activation. For instance, ligand binding leads to methylation (M) of ER, and subsequent formation of the ER-PI3K-Src-focal adhesion kinase (FAK) complex which activates Akt, and ultimately alters gene expression. GF: Growth factor; RE: Response element; VEGF: Vascular endothelial growth factor; IGFR1: Insulin-like growth factor receptor 1; TGFα: Transforming growth factor α.
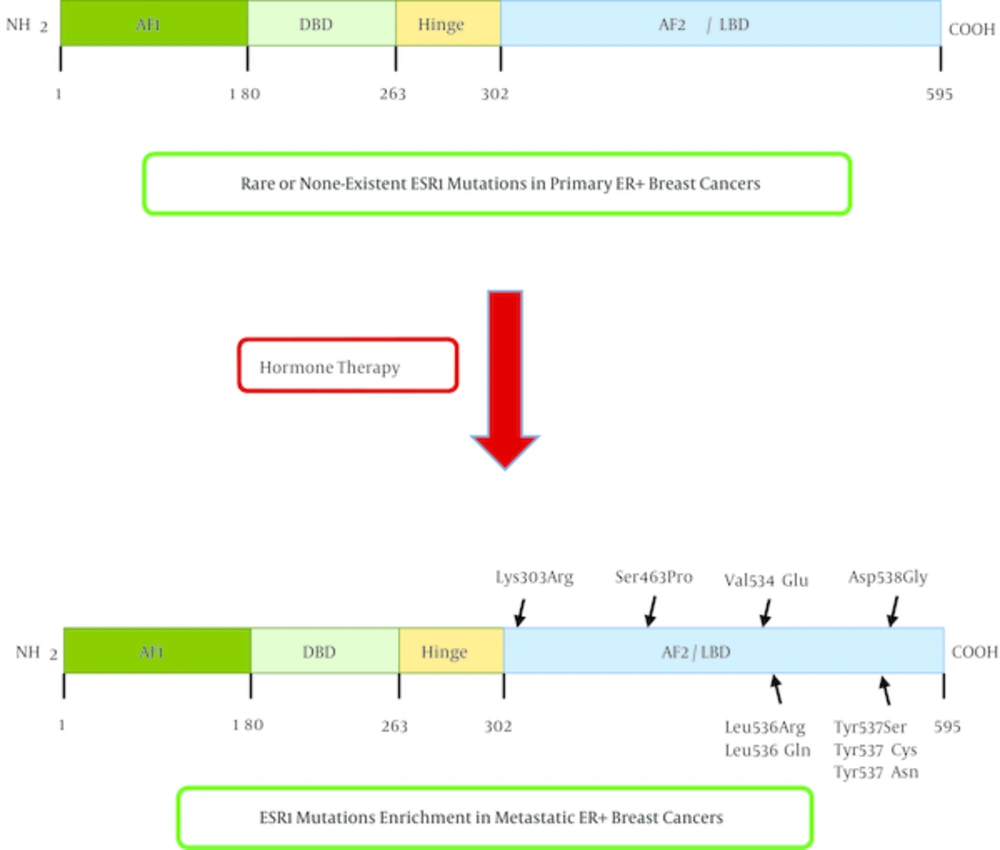
ERα consists of a ligand-independent transactivation domain (AF1), a highly conserved DNA binding domain (DBD) that recognizes estrogen response elements (EREs), a hinge domain, a ligand binding domain (LBD), and a second transactivation domain that is activated after ligand binding (Figure 2) (14). Ligand binding to ERα activates this receptor through phosphorylation, dissociation of chaperonin proteins like heat shock protein 90 and conformational change. Hormone bound ERα then binds to another ERα (dimerization), and the dimer binds to estrogen response elements in the promoter of estrogen responsive genes. Promoter-bound ERα dimers form complexes with co-regulatory proteins such as AIB1, which subsequently result in expression of estrogen responsive genes (Figure 1).
ESR1 mutations are rare in primary ER + breast cancers, and are considerably enriched in metastatic, hormone therapy resistant breast tumors. Almost all of the mutations in the estrogen receptor are localized in the ligand binding domain (LBD) of the estrogen receptor and leads to constitutive activation of the protein. AF1: Activation Function 1; DBD: DNA binding domain; AF2: Activation function 2; LBD: Ligand binding domain.
2. Evidence Acquisition
Using keywords such as “breast cancer”, “estrogen receptor”, “hormone therapy”, “hormone therapy response”, “hormone therapy resistance”, “tamoxifen resistance”, “ESR1 amplification” and “ESR1 mutation”, a thorough PubMed search was conducted to find high quality articles pertaining to the central theme of this study. Articles from the past 20 years were analyzed for pertinent information, and subsequently reviewed by the authors. Articles deemed to provide valuable information to the topic were isolated for further analysis and included in the content of this paper.
3. Results
3.1. ESR1 Mutations in Breast Cancer
Recurrent gain-of-function mutations in several oncogenes and signaling molecules have been reported in various cancers during tumor development and progression. Recently, ESR1 mutation has emerged as a potential mechanism implicated in breast cancer progression and acquired endocrine resistance. The mutations occurring in the ligand binding domain (LBD) of ERα create ligand-independent constitutively active estrogen receptors and maintain ER signaling. The incidence of these mutations in breast cancers has been reported to be as low as less than 1% in primary cancers, but as high as 11% - 55% in metastatic ER + tumors (15-17).
The first report of ESR1 mutation was described in a patient with metastatic breast cancer in 1997 (18). They identified three missense mutations, Ser47Thr, Lys531Glu, and Tyr537Asn after analyzing 30 metastatic breast samples. The patient with Tyr537Asn substitution had been on hormone therapy before recurrence of cancer as bone metastasis.
The Cancer Genome Atlas (TCGA) Network published the result of analysis of 825 primary breast cancer tumors by genomic DNA copy number arrays, DNA methylation, exome sequencing, mRNA arrays, miRNA sequencing and reverse phase protein arrays (19). They were not able to find any ESR1 mutations in this large cohort of breast cancer samples.
Butler et al. performed whole exome sequencing of cfDNA (cell free DNA) and tumor DNA from two patients with metastatic cancer (20). One of the patients was a female with ER + breast cancer and metastasis to bone and liver on anastrozole therapy for 33 months. They identified Asp538Gly mutation in ESR1 gene in both metastatic sites and the cfDNA. This mutation was not present in the primary breast cancer sample.
Merenbakh-Lamin et al. studied 13 tumor samples from ER + metastatic breast cancer patients with deep sequencing of several cancer-related genes (21). All of these patients had failed multiple lines of endocrine therapy. They identified Asp538Gly mutation in ESR1 gene in liver metastasis obtained from 5 patients. This mutation was not detected in the primary tumor of these patients, obtained prior to initiation of any hormone treatment. All 5 patients had received at least two lines of hormone therapy for a long period. In one patient with ERα mutation in liver metastasis, no mutation was found in pulmonary metastasis. This observation may be due to emergence of the ERα mutation in the liver or improved survival of cells harboring this mutation in the liver tissue.
Toy et al. studied 80 tumors from patients with metastatic ER + breast cancer who had received hormone therapy and whose tumors had grown or spread to new sites on therapy (16). They used a targeted approach to characterize genomic alterations including mutations and copy number alterations among 230 genes frequently mutated in cancer by next generation sequencing (NGS). They identified on average 4.3 mutations per tumor with an average coverage depth of ~ 500X. After comparison with the frequency of mutated genes in primary (untreated) breast tumors analyzed by TCGA, they identified mutations in 3 genes (ESR1, RPTOR and ERBB3) at much higher rate in their recurrent tumor samples. In contrast, the prevalence of mutations in TP53, PIK3CA and GATA3 were comparable between the two sets. Amplification of ERBB2, CCND1 or FGFR1 was commonly detected in this recurrent tumor set, but was not observed to be “acquired” in those tumors that information on primary tumor was available. They identified 14 mutations in the ligand binding domain of ESR1. The clinical history of these patients revealed that they had received multiple lines of hormone therapy for an average of 4.9 years. The most frequent mutations involved Tyr537 and Asp538 of the LBD.
Robinson et al. analyzed 11 patients with metastatic ER positive breast cancer (15). They studied a metastatic tumor biopsy with matched normal samples (blood and/or buccal swabs) using whole exome and transcriptome sequencing. The most frequent mutational observation was ESR1 LBD domain mutations in 6 patients. These mutations included p.Leu536Gln, p.Tyr537Ser, p.Asp538Gly, p.Tyr537Ser, p.Asp538Gly and p.Tyr537Ser alterations. All of these patients were treated with anti-estrogens (tamoxifen and/or fulvestrant) and aromatase inhibitors (letrozole, anastrozole, and/or exemestane). Two of the breast cancer patients also had an oophorectomy. For three of the patients, they were able to analyze the primary breast tumors with no evidence of ESR1 mutation, reflecting the acquired nature of these ESR1 mutations after endocrine therapy. ESR1 amplifications were not observed in these cases.
Jeselsohn et al. analyzed 134 ER positive and 115 ER negative breast cancer tumors with next generation sequencing targeting ESR1 and 182 cancer-related genes (22). The ER + samples consisted of 58 primary tumors and 76 metastatic tumors. ER+/HER2+ tumors were excluded from the study, as they are known to be resistant to hormone therapy. They detected 12 somatic mutations in the LBD domain of ERα gene. The mutations included Tyr537Asn, Tyr537Ser, Tyr537Cys, and Asp538Gly. These ERα mutations were detected in none of the primary treatment-naive ER + tumors or ER- tumors. One ERα mutation (Arg503Trp) was detected in a primary ER- tumor, which is not a known somatic variant of ESR1 in the literature. Two instances of ESR1 focal amplification was also observed. The study included 37 matched primary and metastatic tumor pairs. In two of tumor pairs, point mutations were detected in the metastatic samples, but not in the matched pretreatment primary tumor samples. In one patient with ESR1 mutation in a lymph node biopsy after 8 years of tamoxifen treatment, a biopsy from the same biopsy site before initiation of tamoxifen therapy was available with no evidence of ESR1 mutation. In 3 specimens, there was evidence of ESR1 mutations in subclonal cell populations, suggesting clonal selection of endocrine resistant phenotype. After analysis of the other 182 cancer-related genes, they realized that none of the commonly altered genes in ER positive breast cancer was mutually exclusive with ESR1 genomic alterations. Furthermore, of the most frequently altered genes, only ESR1 alterations showed significantly different frequencies across primary and metastatic tumor samples, suggesting a role for aberrant ERα in the development of recurrence.
Niu et al. analyzed 341 advanced breast cancer patients with targeted next generation sequencing (17). The inclusion criterion was a recurrent breast cancer which had not responded to or had progressed on two lines of standard therapy. They did not analyze any primary breast tumors. 10.7% of the samples were local recurrences and 89.3% were from metastatic sites. ESR1 mutations were identified in 27 patients with ER + breast cancer. All of the samples harbored mutations in the ligand binding domain of the ESR1 gene, with the most common mutations affecting tyrosine 537 (60.7%) and aspartic acid 538 (32.1%). They also found ESR1 amplification in one patient.
Sefrioui et al. analyzed 7 metastatic ER+ breast cancer patients previously exposed to aromatase inhibitors (AIs) who had an initial response to AI, but subsequently developed resistance (23). They studied ESR1 mutations in the primary tumor before any treatment, pleural metastasis and plasma samples during disease progression. No mutations were detected in the primary tumors, neither by Sanger sequencing nor by digital PCR. Three different mutations (Asp538Gly*2, Tyr537Asn, and Tyr537Ser) were detected in four metastasis samples by Sanger sequencing. One sample had an uncertain Asp538Gly mutation. Reanalysis of theses samples by digital PCR identified seven ESR1 mutations in 6 samples, with a double mutation detected in one metastasis sample. Analysis of plasma samples for circulating tumor DNA (ctDNA) by digital PCR identified four ESR1 mutations in the patients with ESR1 mutant metastasis tumors by Sanger sequencing (including the uncertain mutation). No ESR1 mutation was detected in the ctDNA of the patient with Tyr537Asn mutation in the metastasis sample.
Takeshita et al. studied 270 primary and 55 metastatic ER + breast tumor samples to detect ESR1 mutations with droplet digital polymerase chain reaction (ddPCR) (24). The developed ddPCR assay could detect the ESR1 mutant molecules with low concentration of 0.25 copies/μL. According to the selected cut-off, ESR1 mutations were identified in 7 (2.5%) of primary tumor samples and 11 (20%) of metastatic samples. Among the ESR1 mutant metastatic tumors, 5 had Tyr537Ser mutation and 4 had Asp538Gly, Tyr537Asn, and Tyr537Cys mutation. In 4 patients, primary breast tumor and metastatic biopsy were available. Four patients had 2 biopsy specimens, one of them performed after treatment failure. Four of these eight patients acquired ESR1 mutation, whereas no ESR1 mutation could be identified at their first biopsy or primary sample.
Wang et al. analyzed 43 primary breast tumors, 12 bone metastases, 38 brain metastases, and 29 cfDNA samples by droplet digital PCR (25). The ESR1 mutations detected were Asp538Gly (n = 13), Tyr537Ser (n = 3), and Tyr537Cys (n = 1). Mutation rates were 7% in primary tumors, 9.1% in bone metastases, 12.5% in brain metastases, and 24.1% in cfDNA samples. Mutation allele frequencies were 0.07% to 0.2% in primary tumors, 1.4% in bone metastases, 34.3% to 44.9% in brain metastases, and 0.2% to 13.7% in cfDNA samples. Two patients had more than 1 ESR1 mutations.
Schiavon et al. studied ESR1 mutations in tumor biopsies and ctDNA samples from 171 women with advanced breast cancer by multiplex digital PCR (26). ESR1 mutations were found exclusively in ER+ breast cancer patients who had previously received aromatase inhibitors (AI). Patients with ESR1 mutations had reduced progression free survival (PFS) on subsequent AI treatment (hazard ratio, 3.1; 95% confidence interval (CI), 1.9 to 23.1; P = 0.0041). ESR1 mutation frequencies differed substantially between patients who received AI in the adjuvant versus metastatic setting (5.8% versus 36.4%, respectively; P = 0.0002). In a patient with available serial sampling, ESR1 mutations became the dominant clone in tumor samples during metastatic AI treatment.
Guttery et al. analyzed ESR1 mutations in cfDNA samples from 48 metastatic ER+ breast cancer patients on systematic therapy by targeted NGS and used ddPCR for ESR1 Asp538Gly mutation as a validation method (27). ESR1 mutations were detected in 9 cfDNA samples. NGS detected 3 mutations in ESR1, including p. Asp538Gly at 46.3%, p. Tyr537Ser at 2.8%, and p.Glu380Gln at 24.4% mutation frequency, respectively. ddPCR was more sensitive than NGS and identified 6 additional ESR1 mutations at a frequency of < 1%. Using a combination of NGS and ddPCR, ESR1 mutations were detected in 18.75% baseline cfDNA samples. In serial blood samples from 11 patients, 2 showed emergence of new ESR1 mutations. In the first patient, ESR1 Asp538Gly mutation was absent in the baseline cfDNA sample, but emerged after 4 months receiving hormone therapy. In the second patient, ESR1 Asp538Gly mutation was present at < 1% mutation frequency in the baseline cfDNA sample, but the frequency increased on hormone treatment. Table 1 lists the published studies on ESR1 mutations in metastatic ER + hormone therapy resistant breast cancers.
| Recurrence Biopsy Site | Detected ERα Mutations | ERα Mutations in the Matched Primary Breast Tumor | Hormone Treatment Before Biopsy | Study |
|---|---|---|---|---|
| Bone metastasis | p.Tyr537Asn | NA | diethylstilbestrol | Zhang et al. (1997) (18) |
| Liver and bone metastasis | p.Asp538Gly | None | Anastrozole | Butler et al. (2012) (20) |
| Liver metastasis | p.Asp538Gly | None | Anastrozole, Fulvestrant, Exemestane, Tamoxifen, Goserelin | Merenbakh-Lamin et al. (2013) (21) |
| Various metastatic sites | p.Ser463Pro | None | AI, SERD, SERM | Toy et al. (2013) (16) |
| p.Val534Glu | ||||
| p.Pro535His | ||||
| p.Leu536Arg | ||||
| p.Tyr537Ser | ||||
| p.Tyr537Asn | ||||
| p.Tyr537Cys | ||||
| p.Asp538Gly | ||||
| Various metastatic sites | p.Leu536Gln | None | Tamoxifen, Letrozole, Fulvestrant, Exemestane, Oophorectomy | Robinson et al. (2013) (15) |
| p.Tyr537Ser | ||||
| p.Asp538Gly | ||||
| Various metastatic sites | p.344insCys | None | Anastrozole, Tamoxifen, Exemestane, Fulvestrant, Letrozole Megestrol, Goserelin, | Jeselsohn et al (2014) (22) |
| p.Glu380Gln | ||||
| p.Tyr537Ser | ||||
| p.Tyr537Cys | ||||
| p.Tyr537Asn | ||||
| p.Asp538Gly | ||||
| Local recurrence or metastasis | p.Val533Met | NA | Fulvestrant, tamoxifen, or exemestane | Niu et al. (2015) (17) |
| p.Tyr537Ser | ||||
| p.Tyr537Cys | ||||
| p.Tyr537Asn | ||||
| p.Asp538Gly | ||||
| p.Leu536_Asp538 > Pro | ||||
| Pleural metastasis / ctDNA | p.Tyr537Asn | None | Anastrozole, tamoxifen, Letrozole, Exemestane | Sefrioui et al. (2015) (23) |
| p.Tyr537Ser | ||||
| p.Asp538Gly | ||||
| Various metastatic sites | p.Tyr537Ser | None | NA | Takeshita et al. (2015) (24) |
| p.Tyr537Asn | ||||
| p.Tyr537Cys | ||||
| p.Asp538Gly | ||||
| Brain and bone metastasis, cfDNA | p.Tyr537Ser | p.Asp538Gly | AI, SERD, SERM | Wang et al. (2015) (25) |
| p.Tyr537Cys | ||||
| p.Asp538Gly | ||||
| Various metastatic sites, ctDNA | p.Leu536Arg | NA | AI, Tamoxifen | Schiavon et al. (2015) (26) |
| p.Tyr537Ser | ||||
| p.Tyr537Asn | ||||
| p.Tyr537Cys | ||||
| p.Asp538Gly | ||||
| cfDNA | p.Glu380Gln | NA | Anastrozole, Tamoxifen, Goserelin, Exemestane, Fulvestrant, Letrozole | Guttery et al. (2015) (27) |
| p.Tyr537Ser | ||||
| p.Asp538Gly |
Abbreviations: AI, aromatase inhibitor; cfDNA: cell free DNA; ctDNA: circulating tumor DNA ; NA, not available; SERD: selective estrogen receptor degrader; SERM, selective estrogen receptor modulator.
Consistent with these studies, mutations in the hotspot region of ER ligand binding domain, especially those affecting residues Tyr537 and Asp538, are gain-of-function mutations that lead to both ligand-independent and enhanced ligand-stimulated ER activity (15, 16, 21). Analysis of the structural consequences of these mutations has revealed that the C-terminal helix of ER Tyr537Ser is stabilized in the agonistic configuration in the absence of its ligand, similar to the estrogen-bound activated wild-type ER. Furthermore, molecular dynamics stimulation of these LBD mutants bound to co-activators confirmed that both of them increased the stability of agonistic conformation of C-terminal helix (16, 21). Fulvestrant and tamoxifen could inhibit the mutated ER at substantially higher doses required to inhibit wild-type ER, illustrating partial resistance to these drugs (15, 16, 21, 22). In addition, mutant ERs are resistant to fulvestrant-induced degradation (22). There is evidence that under hormone depleted conditions, ER-LBD mutants co-precipitate more with the SRC-3 co-activator protein compared to wild-type ER (16). A ligand independent interaction between Asp538Gly and SRC-1 co-activator protein has also been reported (21). Studies to directly analyze the affinity of these ER-LBD mutants for tamoxifen and fulvestrant have not been performed. However, it has been identified that in comparison to wild-type ER, mutant ER show increased co-regulator binding after tamoxifen treatment (28).
Recent studies have shown that breast cancers harboring ER-LBD mutations are relatively resistant to tamoxifen and fulvestrant, but these tumors can be effectively inhibited with higher doses of these agents (15, 16, 21, 22). It is possible that higher doses of tamoxifen or fulvestrant or mutant specific selective estrogen receptor modulators (SERMs) or selective estrogen receptor degraders (SERDs) might benefit patients with ER-LBD mutations. The CONFIRM study demonstrated that higher dose of fulvestrant increased survival of ER positive breast cancer patients without additional significant toxicity; therefore, future studies can determine whether high dose fulvestrant can inhibit these ER-LBD mutant breast cancers (29). Another possibility is to study whether high dose tamoxifen is effective in ER-LBD mutant breast cancer, as acceptable toxicity profile of high dose tamoxifen has been reported in some studies (30, 31). New generation SERMs and SERDs, such as bazedoxifene and ARN-810, are being analyzed in preclinical studies to determine their efficiency in the inhibition of ER-LBD mutants (32, 33). As the activity of mutated ER depends much on the recruitment of co-activators, such as SRC-3, the inhibitors of these co-activators may be able to target these ER mutants (34, 35). Furthermore, because of the constitutive ligand independent activity of ER mutants, it is possible to target the classical ER downstream products such as blocking the cyclin D1 with CDK4/6 inhibitors (36). Implementation of these innovative strategies holds the promise to improve patient outcome.
3.2. ESR1 Amplification in Breast Cancer
Gene amplification, defined as a focal increase in copy number of a DNA region, is an important mechanism for increasing protein expression during tumor development. The extra copies of the amplified DNA may be located in one chromosomal region, visible as homogenously staining regions (HSRs) or dispersed as double minute chromosomes (37).
The first report of ESR1 amplification was described by Holst et al. in 2007 (38). They detected ESR1 amplification in 21% of 2000 breast cancer samples by fluorescent in situ hybridization (FISH) method. ESR1 amplification was significantly correlated with ERα expression (P < 0.0001). In 175 patients who had received adjuvant tamoxifen monotherapy, ESR1 amplification was significantly associated with longer survival (P = 0.023). Adelaide et al. studied 274 breast tumors to detect ESR1 amplification with array comparative hybridization method (aCGH) and were not able to detect any ESR1 amplification in this cohort of patients (39).
Tomita et al. analyzed ESR1 amplification in 133 primary breast cancer samples by FISH (40). 23% of cases showed amplification of ESR1. Patients with ESR1 amplification experienced longer disease free survival. Other groups tried to reproduce these findings, but were not successful. Moelans et al. analyzed the frequency of ESR1 amplification in 135 breast tumors by multiplex ligation-dependent probe amplification (MLPA) and FISH methods. ESR1 amplification was detected in only 2% of samples (41). We analyzed gene amplifications by MLPA and FISH methods in 170 breast cancer tumors from patients who had received tamoxifen (11, 42). ESR1 amplification was detected only in 2% of samples and there was no association between ESR1 amplification and recurrence on tamoxifen.
Ooi et al. studied 106 breast tumors for ESR1 amplification using FISH method (43). They showed that the large clustered signals previously interpreted as ESR1 amplification are sensitive to RNase treatment. Their findings indicate that FISH detects accumulation of ESR1 transcript in the nucleus of breast cancer cells expressing high levels of ERα. Using touch smears from 51 fresh breast tumors, enumeration of FISH signals after RNase treatment revealed ESR1 amplification in 6% of samples.
Moelans et al. analyzed 35 grade 3 breast cancers for ESR1 amplification detection with FISH/ MLPA (44). RNase pretreatment resulted in a higher fraction of cells with point-shaped FISH signals. But FISH with and without RNase pretreatment showed no major differences in ESR1 amplification. These results suggest that reasons other than RNA-related hybridization artifacts account for the different ESR1 amplification reported in different studies. Chen et al. tested 311 breast cancers for ESR1 amplification with FISH and chromogenic in situ hybridization (CISH) methods (45). None of the tumors analyzed showed ESR1 amplification with either method.
In general, ESR1 amplification frequency is highly discordant among studies that used FISH. This variability may be a consequence of different protocols, scoring systems, and FISH analysis artefacts due to accumulation of nuclear ESR1 transcripts in cells with high level ER expression, which could be misinterpreted as ESR1 amplification. More recent studies using NGS have reported that the rate of ESR1 amplification in both primary and metastatic breast tumors is approximately 2% (19), indicating that ERS1 amplification may not have a major role in the development of hormone therapy resistance. Nevertheless, results of a study of neoadjuvant endocrine therapy showed that 6 months after treatment a new amplicon containing ESR1 was acquired (46). Therefore, the clinical significance of ESR1 amplification in early and metastatic breast cancer remains debatable.
4. Conclusions
In summary, ESR1 mutations are considerably enriched in hormone therapy resistant, metastatic ER positive breast cancers and are rare or absent in primary, hormone therapy-naïve breast tumors (Figure 2). On the basis of published studies, the frequencies of ESR1 mutations in ER positive metastatic breast cancer range from 11% to 55% (15, 16, 21, 22). These differences in ESR1 mutation frequency reported by various groups may be due to different patient cohorts and different methodologies of mutation detection with different sensitivities. Innovative platforms that enable the detection of rare subclonal mutations are essential to elucidate ESR1 mutation prevalence. Future larger prospective studies with standardized mutation detection methods may be necessary to determine the true incidence of ESR1 mutations. Furthermore, the causal relationship between emergence of ESR1 mutations and endocrine resistance needs to be established. Detection of such mutations in liquid biopsies might help in early detection of resistant occult micrometastatic residual disease during or after adjuvant hormone therapy.
ESR1 amplification in breast cancer remains a controversial issue. Although Ooi et al. showed that the large clustered ESR1 FISH signals that have been interpreted as amplification, are sensitive to RNase treatment (43), other groups have not been able to replicate their results (44). The combination of intra-tumor heterogeneity regarding ESR1 copy number alterations and low level ESR1 copy number increase may account for these discrepancies.
In conclusion, while numerous unknown issues on the role of ESR1 mutations in advanced breast cancer remain, these new findings will certainly deepen current knowledge on molecular evolution of breast cancer and acquired resistance to hormone therapy. Future studies will disclose the overall contribution of ESR1 genetic aberrations including mutations and amplifications to hormone resistance and their contribution in clinical decision making.