1. Background
Out of the estimated 10 million of human single nucleotide polymorphisms (SNPs) in the human genome, only 3% to 5% are located in the coding regions (1). They are classified into synonymous, (sSNPs), and non-synonymous SNPs, (nsSNPs) (2). nsSNPs that occur in the genes encoding for drug target proteins could affect target protein functions and hence its drug-target interactions. Because of their functional importance, nsSNPs represent the most well understood group of genetic variations (3). The human population is estimated to have 67,000 to 200,000 common nsSNPs (1, 4, 5), and each person is thought to be heterozygous for 24,000 to 40,000 of these variants (1).
Topoisomerase II-α (Top2-α) is an isomer of Top2 enzymes that controls and alters the topologic states of DNA. In humans, this enzyme is encoded by the TOP2A gene localized to chromosome 17 (6). As Top2-α is an important target for many anti-cancer drugs, variations in this enzyme may influence drug responses in different populations. A variety of mutations in this gene have been associated with the development of drug resistance (7-10).
Recently, we computationally examined the mechanisms, by which nsSNP variations in Top2-α could affect its response to Amsacrine and Mitoxantrone as important inhibitors of the enzyme. The results of the study showed that SNP rs267607133 (c.1460G>A, p.Arg487Lys) could significantly affect the position and binding energy of Amsacrine in Top2-α, which could mediate resistance of this polymorphism to Amsacrine (11). Despite the importance of nsSNPs in the response of Top2-α to anti-cancer drugs, allelic frequencies of these variants have not been yet studied. In the present study, the allele frequency of two novel SNPs altering human DNA Top2-α interaction with Amsacrine and Mitoxantrone (11) was surveyed in the Iranian population. These SNPs were non-synonymous mutations leading to changes in the amino acid sequence: rs762022284 (c.1585A>G, p.Lys529Glu) and rs764177670 (c.1589C>T, p.Thr530Met).
2. Methods
2.1. Genotyping of nsSNPs
The study population was composed of 100 unrelated healthy individuals (average 45.4 years) in the Iranian population. The blood samples were obtained from the Isfahan Medical Genetics Center, Isfahan, Islamic Republic of Iran. The blood donors had no history of personal or familial cancer. Genomic DNA was extracted from the leukocytes by a standard salting out procedure (12). The sequence of nsSNPs involved in altered response of Top2-α to its inhibitors (11) was obtained from dbSNP (13). Amplification-refractory mutation system (ARMS)-PCR (14) was carried out for genotyping of SNP variants with specific primers (Table 1). Amplification reactions were performed in a total volume of 25 μL containing 1 µL of genomic DNA (~ 100 ng), 3.0 mM MgCl2, 10X reaction buffer (200 mM Tris-HCl, pH = 8.4, and 500 mM KCl), 1 mM of dNTPs, and 1 U of Taq polymerase. The PCR conditions consisted of one cycle at 95°C for 5 minutes, followed by 35 cycles at 95°C for 1 minute, 59°C for 1 minute, 72°C for 1 minute, and 10°C for 5 minutes. PCR products were separated in a 2% agarose gel, stained with ethidium bromide and visualized on a UV transilluminator. Finally, Sanger sequencing was carried out to verify the genotype of the variants (ABI PRISM 3100, ABI, USA).
| Primer Name | Sequence (5´ → 3´) | PCR AT, °C | Fragments, bp | |
|---|---|---|---|---|
| CF1 | CATTGTAGCCTATTTTAACAGACCT | 464 | ||
| CR1 | CACTTAACCTGTATTTGCAACATAC | |||
| rs762022284 A>G (K529E) | 59 | 270 | ||
| Reverse, A allele | ATTATCTTCCCATAACGAAGCGTATT | |||
| Reverse, G allele | ATTATCTTCCCATAACGAAGCGTATC | |||
| rs764177670 C>T (T530M) | 59 | 273 | ||
| Reverse, C allele | ATCATTATCTTCCCATAACGAATCG | |||
| Reverse, T allele | ATCATTATCTTCCCATAACGAATCA |
Abbreviations: CF, common forward; CR, common reverse; PCR AT, PCR annealing temperature.
2.2. Statistical Analysis
PLINK program (15) was used for the calculation of genotype and allele frequencies, Hardy-Weinberg equilibrium (HWE), linkage disequilibrium (LD), and haplotype frequencies. Gene heterozygosity (He) and PIC (polymorphism information content) values were also calculated with PICcalc program (16).
Genotype and the derived allelic frequencies were estimated based on the observed numbers of the specific alleles from the genotype. The genotype frequency for each polymorphism was, then, tested for deviation from HWE, using a chi-squared goodness-of-fit analysis. He and PIC were calculated according to the methods of Nagy et al. through PICcalc (16). Haplotypes were constructed based on linkage disequilibrium (LD) that was computed for SNP pairwise. Results for LD analysis were considered statistically significant at R2 > 1.
3. Results
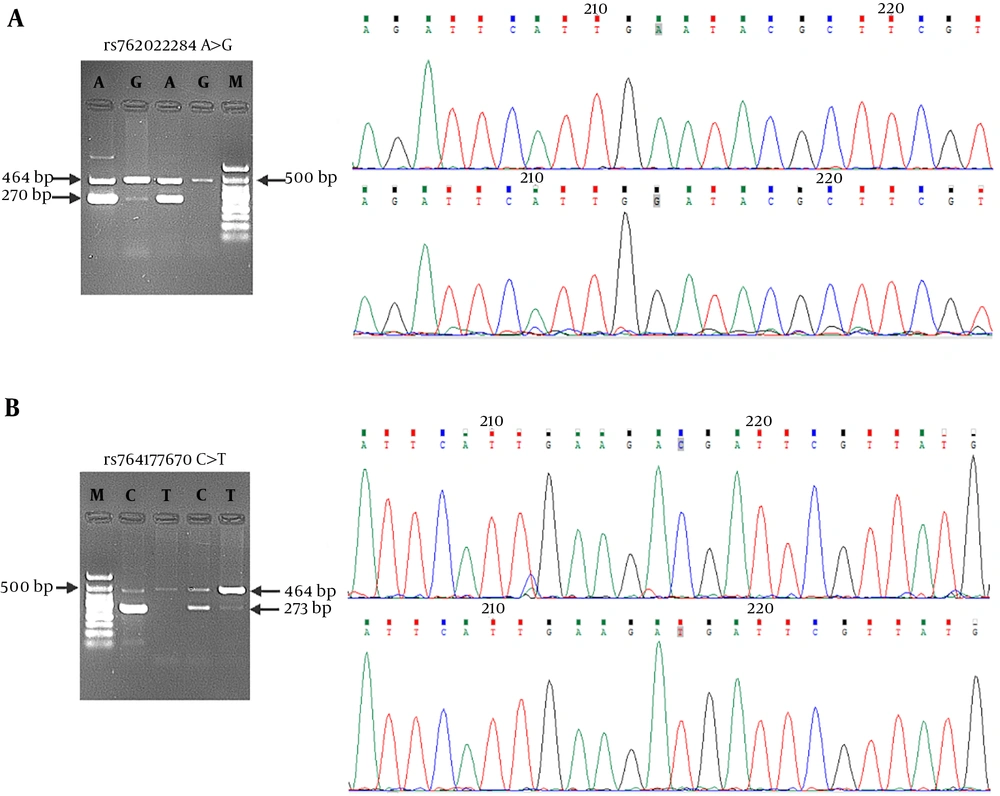
Given the in silico effects of rs762022284 and rs764177670 nsSNPs on Top2-α/drug interaction as in our previous study (12), their genotypes were investigated in the population. The expected genotypes for the selected SNPs were shown in Table 2 and Figure 1, respectively. Genotypic distributions and minor allele frequency (MAF) for genotyped SNPs in individuals were also shown in Table 2. As shown, allele frequency was 0.30 for rs762022284 and 0.05 for rs764177670 variant in this sample. Moreover, minor alleles for both SNPs were found only in the heterozygous state.
| Locus | Genotype Frequency (%) | MAF | HWE P Value | He | PIC | ||
|---|---|---|---|---|---|---|---|
| Male | Female | Total | |||||
| rs762022284 A>G (K529E) | 0.30 | 0.11 | 0.42 | 0.33 | |||
| N | 48 | 36 | 84 | ||||
| AA | 12 (25) | 55.56 (20) | 32 (38.10) | ||||
| AG | 36 (75) | 44.44 (16) | 52 (61.90) | ||||
| GG | 0 | 0 | 0 | ||||
| rs764177670 C>T (T530M) | 0.05 | 1 | 0.095 | 0.09 | |||
| N | 48 | 32 | 80 | ||||
| CC | 48 (100) | 28 (87.5) | 76 (95) | ||||
| CT | 0 | 12.5(4) | 4 (5) | ||||
| TT | 0 | 0 | 0 | ||||
Abbreviations: He, gene heterozygosity; HWE P value, Hardy-Weinberg equilibrium P value; PIC, polymorphism information content; MAF, minor allele frequency.
Genotyping of (A) rs762022284 and (B) rs764177670 in the Iranian population. Genotyping was performed by ARMS-PCR, using specific primers as mentioned in the materials and methods. For each SNP, the expected fragments were shown for homozygous and heterozygous individuals and longer fragments showed internal bonds. The results of Sanger Sequencing of inner bonds and the position of variations (gray squares) were also shown.
HWE P value, gene heterozygosity and polymorphism information content for all the selected SNPs were summarized in Table 2. The data showed that the genotypes for all the SNPs were in Hardy-Weinberg proportions (P > 0.05). As shown in Table 2, the degree of heterozygosity was 0.42 (PIC = 0.33) and 0.095 (PIC = 0.09) for rs762022284 and rs764177670 variants, respectively. Moreover, the data for LD and Haplotype analysis of SNPs, as shown in Table 3, indicated that SNPs rs762022284 with rs764177670 spanned in LD block, suggesting the presence of LD.
| Polymorphisms/Haplotype | Frequency | R-sq |
|---|---|---|
| rs762022284 rs764177670 | 0.059 | |
| GT | 0.026 | |
| AT | 0 | |
| GC | 0.289 | |
| AC | 0.684 |
4. Discussion
SNPs represent the most frequent type of human population DNA variation (17). SNP analysis is used throughout the life sciences including molecular diagnostics, agriculture, food testing, identity testing, pathogen identification, drug discovery and development, and pharmacogenomics (18). One of the main goals of research on SNPs was to understand the correlation of genotype/phenotype of the human genome variations, especially those related to human complex diseases (17). Non-synonymous polymorphisms, nsSNPs, can affect protein function, and are believed to have the largest impact on human health compared with SNPs in other regions of the genome (19). Since Top2-α is an important target for many anti-cancer drugs (20), variations in this enzyme may influence drug responses in different populations. In this study, two nsSNPs rs762022284 (c.1585A>G, p.Lys529Glu) and rs764177670 (c.1589C>T, p.Thr530Met) in TOP2A gene altering drug response of Top2-α (11) were tested in the Iranian population. These SNPs were already investigated by in silico tools in our previous study, showing to affect the activity of Top2-α in response to two important chemotherapy drugs including Amsacrine and Mitoxantrone (12).
Treatment with Top2-α poisons is usually associated with an increased risk of secondary malignancies and cardiotoxicity. Therefore, it is very important to know that whether the drug could have effective treatment function (21, 22). The use of a SNP associated with drug response can be judged in terms of how well it predicts drug response in patients, and the proportions of patients, who will benefit from the test (23). Genotyping of SNPs, which were involved in drug resistance, could provide a cost-effective solution to reduce the side effects of the chemotherapy drugs such as Top2-α inhibitors. Therefore, with this goal, population study of such SNPs would be very important in identifying the effective genotypes involved in drug responses.
In the present study, genotyping of Top2-α SNP variants was performed, using ARMS-PCR (14). As illustrated in Figure 1, the expected alleles were obtained and only heterozygous state was observed for both SNPs. In the human genome, the nsSNPs were enriched in low-frequency alleles compared with the other types of SNPs. On the other hand, studies showed that more than half (59%) of nsSNPs were found at a frequency below 5% (1). There is a minor allele frequency of 0 for rs762022284 in the ExAC and GnomAD databases. Moreover, rs764177670 has a minor allele frequency of 0, 0.00006, and 0.00005 in the ExAC, GnomAD, and TOPMED databases, respectively. As shown in Table 2, allele frequency was 5% for rs764177670 variant and surprisingly it was 0.30 for s762022284, which showed a high frequency for this SNP in the Iranian population.
The heterozygosity of a locus is defined as the probability that an individual is heterozygous for the locus in the population (24). As mentioned above, a high heterozygosity was observed for rs762022284 variant in the Iranian population (with He = 0.42). PIC also showed diversity for rs762022284 in the studied population. PIC refers to the value of a marker for detecting polymorphism within a population, depending on the number of detectable alleles and the distribution of their frequency. This could provide an estimate of the discriminating power of the marker (25).
4.1. Conclusions
In this study, two novel SNPs altering human DNA Top2-α interaction with Amsacrine and Mitoxantrone were genotyped in the Iranian population. The results showed that the allele frequency of rs762022284 was surprisingly high, suggesting that the presence of this SNP in Top2-α could affect cancer therapy with Amsacrine and Mitoxantrone in the Iranian population.