Dear Editor,
Over-activation of Ras signaling pathway is implicated in the pathogenesis of several malignancies including breast cancer. Ras Guanyl nucleotide releasing peptides (RasGRPs) are a group of Ras guanine nucleotide exchange factors (RasGEFs), which participate in the activation of Ras signaling pathway. The RasGRP3 induces the expression and function of H-Ras and R-Ras proteins and plays oncogenic roles in numerous malignancies (1). Over-expression of RasGRP3 protein has been demonstrated in human breast tumor tissue samples compared with normal breast samples. Its silencing in breast cancer cells has diminished cell proliferation, triggered apoptosis in MCF7 cells, and made T-47D cells responsive to Tamoxifen and trastuzumab (1). On the other hand, RASGRP3 has been predicted to be a target of miR-100 regulatory function (2) a miRNA, whose participation in breast cancer pathogenesis has been highlighted (3). In order to find the clinical relevance of RASGRP3 expression in breast cancer patients, we evaluated RASGRP3 expression in 50 invasive ductal carcinoma samples compared with the corresponding adjacent non-cancerous tissues (ANCTs) by means of quantitative real time PCR after receiving the approval of Ethics Committee of Shahid Beheshti University of Medical Sciences (IR.SBMU. MSP.REC.1396.451) and obtaining informed consents from all patients. All patients were sporadic cases of breast cancer and had no history of radio/chemotherapy. The nucleotide sequences of primers are as follow:
RASGRP3-F: CACGCCTCAAAGAGACCCAT
RASGRP3-R: AATTGCCGTTGGAGGAGACC
B2M-F: AGATGAGTATGCCTGCCGTG
B2M-R: GCGGCATCTTCAAACCTCCA
B2M gene was used as normalizer. The association between RASGRP3 expression and clinicopathological data was assessed, using Chi-square test. The correlation between the relative expressions of RASGRP3 and miR-100 was evaluated, using Spearman's rank correlation coefficient. The level of significance was set at P < 0.05.
In addition, through the in silico application of miRNA target prediction tools, we searched for miRNAs that target RASGRP3 mRNA. As miR-100 was one of the miRNAs predicted to affect RASGRP3 mRNA with strong evidences, we subsequently the extracted data of miR-100 expression analysis in the same cohort of patients (unpublished data) and assessed the correlation between the expression levels of RASGRP3 and miR-100.
Next, we used the gene expression omnibus (GEO) source at the National Center for Biotechnology Information (NCBI) archives (4) to obtain GSE59246 and GSE59247 microarray gene expression data, which included mRNA and miRNA expression profiles of normal subjects and patients with breast cancer, respectively. Table 1 shows the characteristics of microarray gene expression datasets.
| GEO Number | Year | Author | Paper Title | Samples | Chip Type |
|---|---|---|---|---|---|
| GSE59246 | 2016 | Lesurf R | Molecular features of subtype-specific progression from ductal carcinoma in situ to invasive breast cancer (gene expression data) | Normal breast tissues: n = 3, tumor tissues: n = 102 | Agilent-028004 SurePrint G3 Human GE 8x60K Microarray |
| GSE59247 | 2016 | Lesurf R | Molecular features of subtype-specific progression from ductal carcinoma in situ to invasive breast cancer (miRNA data) | Normal breast tissues: n = 3, tumor tissues: n = 45 | Agilent-031181 Unrestricted-Human-miRNA-V16.0-Microarray 030840 |
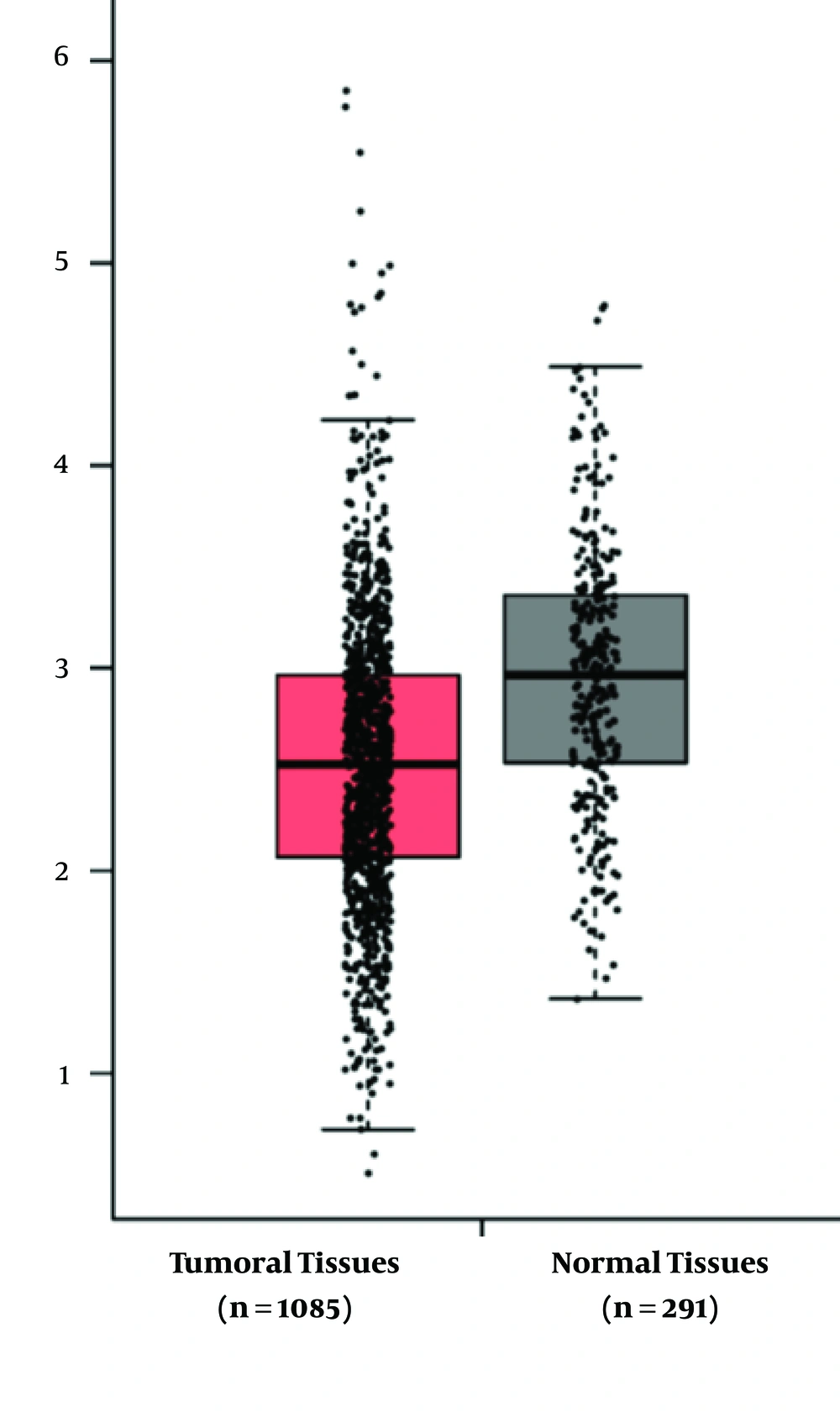
To confirm these results, we subsequently used analysis of RNA Sequencing data, using gene expression profiling interactive analysis (GEPIA) tool (5) to get the RNA sequencing expression data acquired from the TCGA and the GTEx projects.
RASGRP3 expression was not significantly different between tumor samples and ANCTs (tumor vs. ANCT: 1.07 ± 0.9 vs. 0.29 ± 1.1, P = 0.21). Besides, no correlation has been detected between the expression levels of RASGRP3 and miR-100 (r = 0.101, P > 0.05). The analysis of association between RASGRP3 expression levels and patients' clinical and demographic data revealed no significant association between its expression and such data including tumor stage and grade, estrogen, and progesterone receptors status or HER2 expression levels (Table 2). We compared RASGRP3 expression between patients with breast cancer and normal samples, using GEO2R web tool (https://www.ncbi.nlm.nih.gov/geo/info/geo2r.html). Furthermore, using these 2 microarray datasets, we assessed the correlations between the expression levels of RASGRP3 and miR-100. We did not detect either significant difference in RASGRP3 expression between cancerous and normal samples (log Fold Change = -0.095, P = 0.06) or any correlation between the expression levels of the mentioned genes. The analysis of data obtained from GEPIA tool confirmed the above-mentioned data regarding the similar expression of RASGRP3 between cancerous and normal tissues (Figure 1).
| Features | No. (%) | RASGRP3 High Expression Cases (No = 24) | P Value |
|---|---|---|---|
| Age | NS | ||
| ≤ 55 | 30 (60) | 17 | |
| > 55 | 20 (40) | 7 | |
| TNM stage | NS | ||
| Low | 32 (64) | 17 | |
| High | 18 (36) | 7 | |
| Tumor grade | NS | ||
| Low | 34 (68) | 11 | |
| High | 16 (32) | 13 | |
| Tumor size | NS | ||
| > 2 | 9 (18) | 3 | |
| ≤ 2 | 41 (82) | 21 | |
| Lymphatic metastasis | NS | ||
| + | 8 (16) | 5 | |
| - | 14 (84) | 4 | |
| History of abortion | NS | ||
| + | 10 (20) | 5 | |
| - | 40 (80) | 19 | |
| ER | NS | ||
| + | 40 (80) | 18 | |
| - | 10 (20) | 6 | |
| PR | NS | ||
| + | 41 (82) | 16 | |
| - | 9 (18) | 8 | |
| Her2/neu | NS | ||
| + | 37 (74) | 20 | |
| - | 13 (26) | 4 | |
| Ki67 | NS | ||
| + | 45 (90) | 20 | |
| - | 5 (10) | 4 |
Abbreviation: NS, not significant.
Previous reports have demonstrated the contribution of RASGRP3 in the pathogenesis of breast (1) and prostate cancers (6). In addition, RASGRP3 has been among the shared targets of miR-92ab and miR-100 within β4 integrin-regulated mRNAs in breast cancer, which have been recognized through the assessment of published Affymetrix GeneChip data (2). However, we could not find any expression change between breast cancer tissues and non-cancerous tissues by means of real time PCR or the assessment of published microarray datasets or RNA sequencing data. Considering the role of high throughput expression analyses in identification of cancer biomarkers and therapeutic targets, the assessment of these available sources has been suggested as an economical and efficient strategy to select possible biomarkers or therapeutic targets. However, due to technical pitfalls in these strategies, the obtained results should be confirmed by other techniques such as real time quantitative PCR. The inconsistencies between the results of the previous studies and our data regarding the expression of RASGRP3 might be explained by the heterogeneity of breast cancer or the possible presence of ethnic-based gene signatures.
On the other hand, despite the results of the in silico prediction tools regarding the regulatory role of miR-100 on RASGRP3, we could not detect any correlation between the expression levels of these two genes in the assessed samples, which might be due to the presence of complex interactive networks in the context of cancer. Future in vitro functional studies are needed to evaluate the regulatory role of miR-100 on RASGRP3 expression.