1. Background
As the most frequently diagnosed type of cancer, breast cancer among women can be classified based on expression patterns to luminal A, luminal B, Her2+, and triple negative (TN) subtypes, which are correlated with patients’ survival and prognosis (1). Several researches have focused on the exploration of gene expression patterns among these subtypes to find a biologically relevant biomarker in breast cancer (2-5). The expression pattern analysis of non-coding RNAs has also been the focus of researchers (6, 7). Pathway-based expression analysis has further been suggested as a more systematic strategy for biomarker discovery (8, 9). MicroRNAs (miRNAs), as the major regulators of gene expression, and pathway regulation are thought to participate in tumorigenesis process by changing the expression of several mRNA coding genes, non-coding RNA as well as cancer-related signaling pathways. MiR-206 is among miRNAs, whose role has been highlighted as a tumor suppressor. It has been the first miRNA detected in breast cancer. A miR-206-binding site has been found within the 3’-untranslated region (3’UTR) of estrogen receptor (ER)-α. ER negative breast cancer cells such as MDA-MB-231 cells have higher expression of this miRNA compared with ER positive cells such as MCF-7 cells. Notably, the forced over-expression of this miRNA in triple negative breast cancer cells has diminished their metastatic potential and decreased the expression levels of matrix metalloproteinase while increased expression of breast cancer metastatic suppressor (BRMS)-1. Functional studies have shown Cx43 to be a target of miR-206. Expression analysis in human breast cancer specimens has demonstrated the associations between miR-206 levels and both lymph node status and Cx43 expression. While miR-206 significantly decreased the proliferation and metastatic potential of cancer cells, it could not suppress tumor initiation in a mouse xenograft model due to the contraindicatory roles of Cx43 during tumorigenesis (10). Its down-regulation in breast cancer tissues has been associated with increased tumor size and advanced tumor stage. Up-regulation of miR-206 in MCF-7 breast cancer cells prevented cell proliferation and cell growth by the inhibition of the G1/S transition. MiR-206 has an inhibitory effect on the expression of cyclinD2 at both the transcript and protein levels. Taken together, miR-206 has been recognized as a critical tumor suppressor gene in breast tissues, a probable prognostic biomarker, or a beneficial target for the treatment of patients with breast cancer (11).
Based on the complex interaction network between miRNAs and mRNAs, which is implicated in physiologic processes and cancer, several studies aimed at predicting the miRNA targets. However, the valid prediction of miRNA targets is still problematic. Merging experimental and computational tools for decoding miRNA functions and targets has been suggested as an attractive approach in this regard (12). Consequently, in the present study, we have suggested a bioinformatics approach for the identification of miRNA-206 target genes implicated in breast cancer based on miRNA target prediction tools and microarray data analysis. This miRNA has been chosen based on its evident role in the suppression of breast cancer and its contribution in the regulation of tumor-associated pathways.
2. Methods
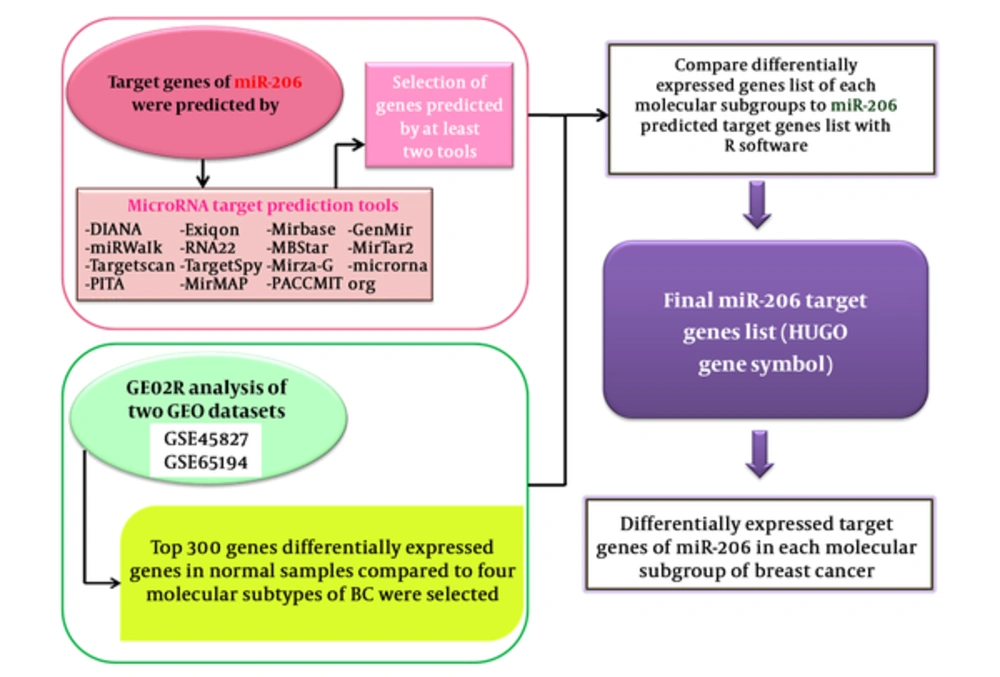
The workflow of data analysis is demonstrated in Figure 1.
2.1. Breast Cancer Expression Data
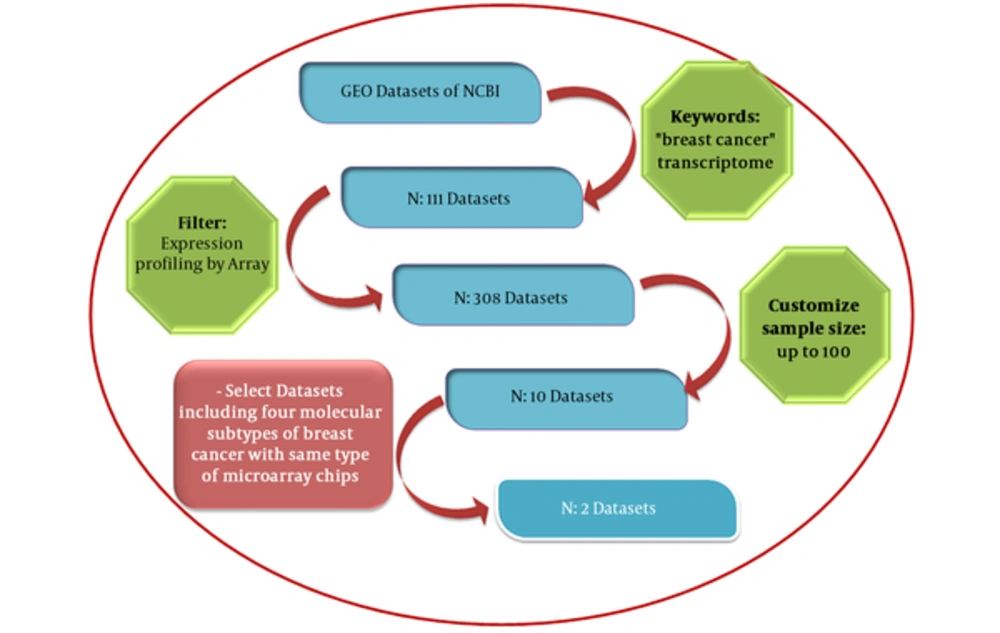
mRNA expression profiles for 285 breast cancer samples and 22 normal tissues were collected from gene expression omnibus (GEO) repository as series GSE65194 and GSE45827. The GEO database launched in 2000 by the national center for biotechnology information (NCBI) incorporates gene expression data provided from microarray technology (13). By entering “breast cancer” and “transcriptome” key words and choosing “Expression profiling by array” as the filter and the minimum sample size of 100 for datasets, 10 datasets were retrieved. GSE45827 and GSE65194 datasets with similar array platforms (GPL570) and inclusion of 4 molecular subtypes of breast cancer (luminal A, luminal B, Her2+ and TN) have been selected for further analyses. The pipeline applied for the selection of these datasets is demonstrated in Figure 2.
2.2. Prediction of miRNA-206 Targets from miRNA Prediction Databases
In this study, systematic miRNA-206 target search was carried out on 14 prediction databases (Mirwalk, MirMAP, Mirbase, PITA, Microrna.org conserved, RNA22, MBStar, DIANA, MirTarbase, mirDIP, MirTar2, Mirza G, PACCMIT, Targetscan, TargetSpy) (Table 1).
| Target Prediction Tool | Organism | Features of Tool | Website |
|---|---|---|---|
| DIANA | Any | Conservation, seed match, and free energy | http://www.microrna.gr/microT-CDS |
| miRWalk | Human, mouse, and rat | Conservation, seed match and free energy | http://mirwalk.uni-hd.de/ |
| Target scan | Human, mouse, Fly, Fish, and Worm | Conservation, seed match | http://www.targetscan.org/ |
| PITA | Human, mouse, Fly, and Worm | Conservation, seed match, free energy, and site accessibility | http://genie.weizmann.ac.il/pubs/mir07/ |
| Exiqon | Human, mouse, and rat | Conservation, seed match, and Target-Site Abundance | https://www.exiqon.com/miRSearch |
| RNA22 | Human, Fruit Fly, Mouse, and Worm | Seed match and free energy | https://cm.jefferson.edu/rna22/ |
| TargetSpy | Human, mouse, rat, Fruit Fly, and Chicken | Seed match and site accessibility | https://omictools.com/targetspy-tool |
| MirMAP | Human, Chimpanzee, Mouse, Rat, Cow, Chicken, Zebrafish, and Opossum | Conservation, seed match, and free energy | http://mirmap.ezlab.org |
| Mirbase | Human, mouse, rat, worm, and fly | Seed match and Target-Site Abundance | http://microrna.sanger.ac.uk |
| MBStar | Human | Seed match and free energy | http://www.isical.ac.in/~bioinfo_miu/MBStar30 |
| Mirza-G | Human | Conservation, seed match, and site accessibility | https://omictools.com/mirza-genome-wide-tool |
| PACCMIT | Any | Conservation, seed match, and site accessibility | http://lcpt.epfl.ch |
| MirTar2 | Human, mouse, rat, dog, and chicken | Conservation, seed match, free energy, and site accessibility | http://mirdb.org |
| Microrna. org | Human, mouse, Fruit Fly, and rat | Conservation, seed match, free energy | http://www.microrna.org |
Summary of miRNA Prediction Tools Used in the Present Study
The fundamental criteria used in some of these prediction databases (for instance TargetScan and DIANA) are complementarity between miRNA and mRNA in seed regions, folding free energy for the miRNA-mRNA duplex, and evolutionary conservation (14). In order to increase the sensitivity of our approach, we chose the mRNAs predicted as miR-206 target by at least 2 databases out of the 15 databases. A list of candidate mRNAs was selected.
2.3. Experimentally Validated miR-206 Target Genes
Due to the drawbacks associated with the predictions of miRNA target sites with the bioinformatics tools (15), in order to find functionally relevant targets in breast cancer, we searched the literature to find functional studies for the identification of miR-206 targets in breast cancer as well as all types of cancer.
2.4. Detection of Differentially Expressed Genes in Breast Cancer Subtypes
We compared the expression profile of 4 breast cancer subtype samples with normal samples by GEO2R web tool (https://www.ncbi.nlm.nih.gov/geo/info/geo2r.html). Based on Log2-fold change between 2 experimental conditions (LogFC) and adjusted P values calculated by the software, 300 genes have been chosen in each subtype with the highest differential expression among tumoral and normal samples.
2.5. Identification of miR-206 Targets Among Differentially Expressed Genes in Breast Cancer Subtypes
The R statistical program (16) was applied to find miR-206 target genes, which are differentially expressed in each cancer subtype compared with normal samples.
2.6. Enrichment Annotation Analysis and Network Construction
FunRich analysis tool version 3 (17) was used for the network construction and the enrichment annotation step. This software depicts a schematic clusterization of the gene list with pathway annotations.
3. Results
3.1. Experimentally Validated miR-206 Target Genes
With the purpose of assessment of the sensitivity of miRNA target prediction tools, we compared the list of experimentally validated miR-206 target genes with those predicted by computational algorithms. The lists of experimentally validated miR-206 target genes in all cancer types and breast cancer as well as the number of bioinformatics tools predicted each gene are presented in Tables 2 and 3, respectively. CCND1 and CCND2 were predicted as targets of miR-206 by a high proportion of tools and were also experimentally validated to be targets of this miRNA in diverse cancer types.
| Target Gene | Number of Tools Predicted the Gene as miR-206 Target | Validation Technique | Disease | Reference |
|---|---|---|---|---|
| CCND2 | 10 tools | Reporter assay | Gastric cancer | (18) |
| MET | 8 tools | Reporter assay, Western blot, qPCR | Rhabdomyosarcoma | (19) |
| GPD2 | 8 tools | Reporter assay, Western blot | Many tumor cell lines | (20) |
| G6PD | 8 tools | Reporter assay, Western blot | Many tumor cell lines | (20) |
| CCND1 | 6 tools | Reporter assay, Western blot | Different cancer cell lines | (21) |
| CDK4 | 5 tools | Reporter assay | Melanoma | (22) |
| PGD | 5 tools | Reporter assay, Western blot | Many tumor cell lines | (20) |
| TKT | 3 tools | Reporter assay, Western blot | Many tumor cell lines | (20) |
| EGFR | 3 tools | Reporter assay, Western blot, qPCR | Lung Squamous Cell Carcinoma | (23) |
Experimentally Validated Target Genes of miR-206 in All Cancers
| Target Gene | Number of Tools Predicted the Gene as miR-206 Target | Validation Technique | Reference |
|---|---|---|---|
| ESR1 | 10 tools | Reporter assay, Western blot, qPCR | (24) |
| CCND2 | 10 tools | Reporter assay | (11) |
| TWF1 | 10 tools | Reporter assay, qPCR and Western blot | (25) |
| SPRED1 | 10 tools | Reporter assay | (26) |
| VEGF | 8 tools | Reporter assay | (27) |
| RASA1 | 8 tools | Reporter assay | (26) |
| NRP1 | 8 tools | Reporter assay | (28) |
| SMAD2 | 8 tools | Reporter assay | (28) |
| MAP3K13 | 6 tools | Reporter assay, qPCR | (29) |
| CCND1 | 6 tools | Reporter assay | (30) |
| Ncl (Nucleolin) | 6 tools | Reporter assay, qPCR and Western blot | (31) |
| Tbx3 | 6 tools | Reporter assay | (32) |
| PDCD4 | 4 tools | Reporter assay | (33) |
| CORO1C | 4 tools | Reporter assay, qPCR and Western blot | (34) |
| PFKFB3 | 3 tools | Reporter assay, qPCR and Western blot | (35) |
| Connexin43 | TargetScan and PicTar | Reporter assay and qPCR | (10) |
Experimentally Validated Target Genes of miR-206 in Breast Cancer
3.2. miR-206 Target Genes Implicated in Breast Cancer
Based on the proposed approach of combining microarray data analysis and bioinformatics prediction tools, we identified miR-206 target genes, which are differentially expressed in breast cancer tissues compared with normal tissues and are possibly implicated in breast cancer (Table 4). The high degrees of similarity in expression profile were observed for SH3GL3, KANK3, and CWF19L2 (overexpressed in all subtypes) and TPM3 (down-regulated in all subtypes). However, some other genes had a subtype specific expression profile. For instance, WDR48 and PTPRS were down-regulated and up-regulated only in luminal B subtype respectively.
| Gene Symbol | Gene Description | HER2 | Triple Negative | Luminal A | Luminal B | Number of Tools Predicted the Gene as miR-206 Target |
|---|---|---|---|---|---|---|
| SH3GL3 | SH3 domain containing GRB2 like 3, endophilin A3 | * | * | * | * | 6 tools |
| PNP | Purine nucleoside phosphorylase | # | # | # | 6 tools | |
| CORO1C | Coronin 1C | # | 6 tools | |||
| WDR48 | WD repeat domain 48 | # | 6 tools | |||
| PTPRS | Protein tyrosine phosphatase, receptor type S | * | 6 tools | |||
| SULF1 | Sulfates 1 | # | # | # | 5 tools | |
| ATP6V1A | ATPase H+ transporting V1 subunit A | # | # | # | 5 tools | |
| YWHAZ | Tyrosine 3-monooxygenase activation protein zeta | # | # | 5 tools | ||
| FN1 | Fibronectin 1 | # | # | # | 5 tools | |
| KIF2A | Kinesin family member 2A | # | 5 tools | |||
| RAB5A | Member RAS oncogene family | # | 5 tools | |||
| UBE2H | Ubiquitin conjugating enzyme E2 H | # | # | # | 4 tools | |
| TPM3 | Tropomyosin 3 | # | # | # | # | 4 tools |
| KDELR2 | KDEL endoplasmic reticulum protein retention receptor2 | # | # | 4 tools | ||
| SFRP1 | Secreted frizzled related protein 1 | * | 4 tools | |||
| TWF1 | Twinfilin actin binding protein 1 | # | 4 tools | |||
| FPGT | Fucose-1-phosphate guanylyltransferase | # | 4 tools | |||
| SRSF1 | Serine and arginine rich splicing factor 1 | # | 4 tools | |||
| GMFB | Glia maturation factor beta | # | # | # | 3 tools | |
| KANK3 | KN motif and ankyrin repeat domains 3 | * | * | * | * | 3 tools |
| CALU | Calcium-binding protein | * | * | * | 3 tools | |
| HOXA5 | Homeobox A5 | * | * | 3 tools | ||
| VMP1 | Vacuole membrane protein 1 | # | 3 tools | |||
| TRIM59 | Tripartite motif containing 59 | # | # | # | 3 tools | |
| SRPK2 | SRSF protein kinase 2 | # | # | 3 tools | ||
| PGM5 | Phosphoglucomutase 5 | * | * | * | 3 tools | |
| MAPK1 | Mitogen-activated protein kinase 1 | # | # | # | 3 tools | |
| ACTR3 | ARP3 actin related protein 3 homolog | # | # | # | 3 tools | |
| CWF19L2 | CWF19 like 2, cell cycle control | * | * | * | * | 3 tools |
| NCBP1 | Nuclear cap binding protein subunit 1 | # | 3 tools | |||
| SRSF3 | Serine and arginine rich splicing factor 3 | # | # | 3 tools | ||
| INMT | Indolethylamine N-methyltransferase | * | * | 3 tools | ||
| SPTBN1 | Spectrin beta, non-erythrocytic 1 | * | 3 tools | |||
| ANP32E | Acidic nuclear phosphoprotein 32 family member E | # | 3 tools | |||
| SDPR | Caveolae associated protein 2 | * | 3 tools | |||
| AMT | Aminomethyltransferase | * | 3 tools | |||
| TBC1D9 | TBC1 domain family member 9 | # | 3 tools | |||
| DDX3X | DEAD-box helicase 3, X-linked | # | 3 tools | |||
| PREX1 | Phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange factor 1 | # | 3 tools | |||
| PCDH19 | Protocadherin 19 | * | 3 tools | |||
| ITIH5 | Inter-alpha-trypsin inhibitor heavy chain family 5 | * | * | 3 tools | ||
| MAPRE1 | Microtubule associated protein RP/EB family member 1 | # | 3 tools | |||
| PRPF4B | Pre-mRNA processing factor 4B | # | # | # | 3 tools | |
| GABRP | Gamma-aminobutyric acid type A receptor pi subunit | * | 3 tools | |||
| LMOD1 | Leiomodin 1 | * | * | 3 tools | ||
| POLR3K | RNA polymerase III subunit K | # | # | 3 tools | ||
| UCHL5 | Ubiquitin C-terminal hydrolase L5 | # | 3 tools | |||
| CA12 | Carbonic anhydrase 12 | # | 3 tools | |||
| EDEM3 | ER degradation enhancing alpha-mannosidase like pro3 | # | 3 tools | |||
| WISP1 | WNT1 inducible signaling pathway protein 1 | # | 3 tools | |||
| RBM47 | RNA binding motif protein 47 | # | 3 tools | |||
| FBXO22 | F-box protein 22 | # | 3 tools | |||
| ARIH1 | Ariadne RBR E3 ubiquitin protein ligase 1 | # | 3 tools | |||
| ZNF146 | Zinc finger protein 146 | # | 3 tools | |||
| ARF4 | ADP ribosylation factor 4 | # | 3 tools | |||
| IKBKB | Inhibitor of nuclear factor kappa B kinase subunit beta | # | 3 tools | |||
| RAB18 | RAB18, member RAS oncogene family | # | 3 tools | |||
| NIPBL | NIPBL, cohesin loading factor | * | 3 tools | |||
| SS18 | nBAF chromatin remodeling complex subunit | # | 3 tools | |||
| ZNF326 | Zinc finger protein 326 | # | 3 tools |
MiR-206 Predicted Target Genes with Differential Expression in Breast Cancer Subtypes Based on the Log2-Transformed Fold Changea
3.3. Network Construction
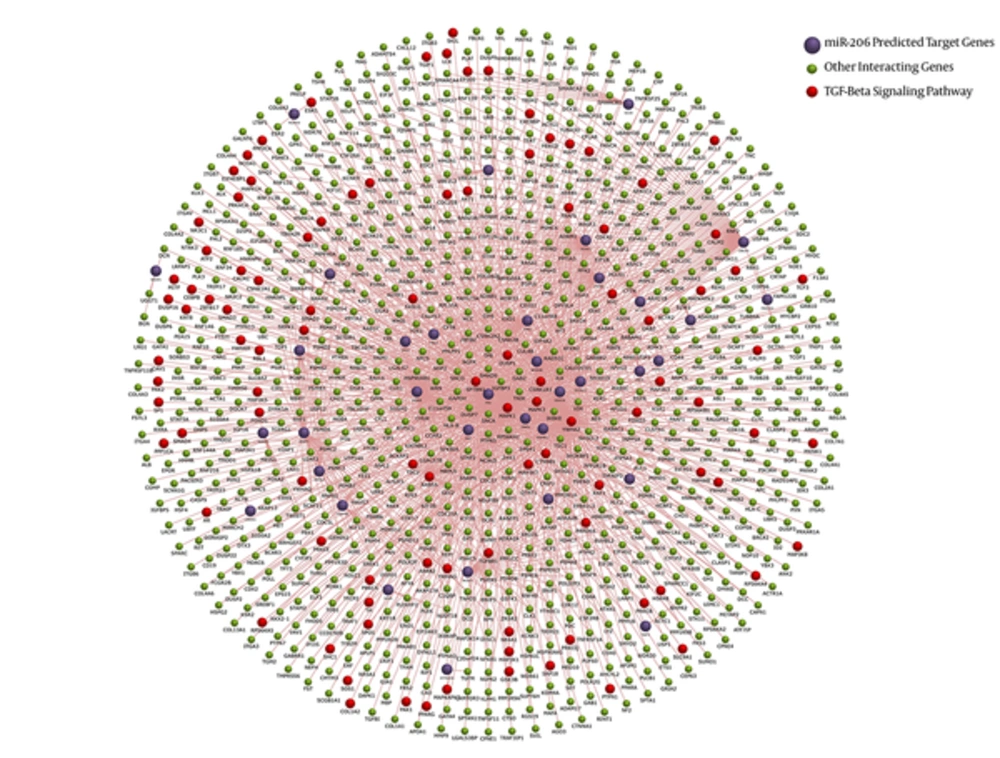
FunRich tool provided both functional enrichment and interaction network for miR-206 and associated mRNAs. The final dataset obtained from the combinatory approach was analyzed against 3 different background databases, namely FunRich, UniProt, and Custom. Figure 3 demonstrates the interaction diagram. Interaction network provided by this tool shows that miR-206 is involved in fundamental pathways in breast cancer. The most enriched one was TGF-beta pathway.
4. Discussion
The expression pattern analysis of distinct subtypes of breast cancer might be helpful in the identification of specific biomarkers as well as the development of personalized treatment modalities (36, 37). The role of miRNAs and their putative targets might be different in each cancer type or even among cancer subtypes. In the present study, we proposed a computational method for the identification of miR-206 targets in distinct breast cancer subtypes. Our suggested approach might guide researchers to arrange their hypotheses of miRNA function on a certain group of microarray samples to choose the candidate mRNAs and miRNAs prior to laboratory experiment. Such approach would also help in the identification of the role of predicted targets as tumor suppressor or oncogenes in signaling pathways. In the final list provided by the proposed approach, there are some genes with expression change in all breast cancer subtypes as well as some genes with alterations in a certain subtype. Such data would help researchers to choose the more suitable cell line for the validation of miR-206 targets.
MiR-206 targets have been validated in experimental studies. For instance, in ER positive MCF-7 cell line, luciferase assays showed that miR-206 decreases cyclinD2 expression by targeting 2 binding sites in the 3’-UTR of cyclinD2 mRNA. The results have been confirmed by qRT-PCR and Western blot assays (11). Another study in the same cell line has shown miR-201 inhibitory effect on the expression of transforming growth factor (TGF)-β, neuropilin-1 (NRP1), and SMAD2, which participate in migration, invasion, and EMT in these cells (28). In TN breast cancer cell line, miR-206 up-regulation has decreased the expression of matrix metalloproteinase (MMP)-2, MMP-9 and increased the expression level of breast cancer metastatic suppressor (BRMS)-1. However, luciferase assays showed only GJA1 (Cx43) as a target of miR-206 (10). Considering the complex interaction network between certain miRNA and numerous mRNAs as well as among various mRNAs, the identification of the net target of each miRNA is of practical significance. Currently, no high-throughput and low-cost miRNA target screening method is available. Although a number of computational methods based on sequence complementarity of the miRNA and the mRNAs have been proposed, the predicted interactions using these computational techniques are unreliable according to high false positive rates. Incorporation of the expression values of miRNAs and mRNAs (and/or proteins) has been suggested as a method to improve the results of sequence-based predictions (12). Our proposed approach provides a tool for combination of miRNA target prediction with mRNA expression data. Although this method is not claimed to be perfect, the suggested list of targets produced by this method is anticipated to significantly lessen laboratory experimental load and the number of interactions to be validated. Homogeneity in experimental conditions should be maintained in our proposed approach. Considering the more significant role of other factors such as transcription factors in regulation of gene expression compared with miRNAs role, heterogeneity of information sources is regarded as a limitation in such approaches (12).
In conclusion, our analysis demonstrated novel mRNA candidates as probable miR-206 targets, which possibly participate in the pathogenesis of breast cancer. The gene list provided by our proposed approach contains some genes previously validated as miR-206 targets in breast cancer (CORO1C, TWF1). However, many of the experimentally validated miR-206 targets are not among gene list provided by our approach. The main cause of such discrepancy is that in our proposed approach, we included genes with high differential expression between tumor tissues and normal samples to find the most biologically relevant targets in breast cancer. However, in the experimental validation of miRNAs targets, the level of expression change is much lower than our defined level in differential expression analyses. For instance, in Yin et al.’s study, the enforced expression of miR-206 resulted in about 50% reduction in the expression of NRP1 and SMAD2 (28). Fu et al. (10) observed an approximately similar reduction in Cx43 levels following the over-expression of miR-206. However, based on our proposed approach, we considered 300 genes with the most differentially expression between tumoral and normal samples, which had at least 10 fold change. Such threshold definition is anticipated to cover the most biologically relevant mRNA targets. A possible limitation of our approach is that by using microarray expression data, miRNA targets can only be detected if the mRNA is degraded by the miRNA. Therefore, our approach may leave out some targets that are influenced at the translation level.