Wilms tumor (WT; OMIM 194070) is a typical embryonic tumor originated from the developmental kidney (1). Mutations in the WT1 gene located in 11p13 have been associated with this malignancy. WT1 gene has 10 exons. The first six exons code for a proline/glutamine-rich transcriptional-regulation region and others code for the four zinc fingers of the DNA-binding domain all implicated either in suppression or in activation of transcription (2). While WT1 gene mutations have been detected in less than 25% of sporadic WTs (3), WT1 abnormalities have been reported in 17% - 81% of hereditary bilateral WTs in different populations (1). There is limited data regarding the inheritance of WT1 mutations in familial cases of WT which have been obtained from the only 13 hereditary WT families described with WT1 mutations (1). WT1 mutations have also been described in juvenile granulosa-cell tumor, non-asbestos-related mesothelioma (4), desmoplastic small round cell tumor (5), and acute myeloid leukemia (6). Although WT1 mutations have been reported in a significant number of cytogenetically normal acute myeloblastic leukemia patients in Iran (6), to the best of our knowledge, there is no report about the prevalence of these mutations in Iranian patients suffering from WT.
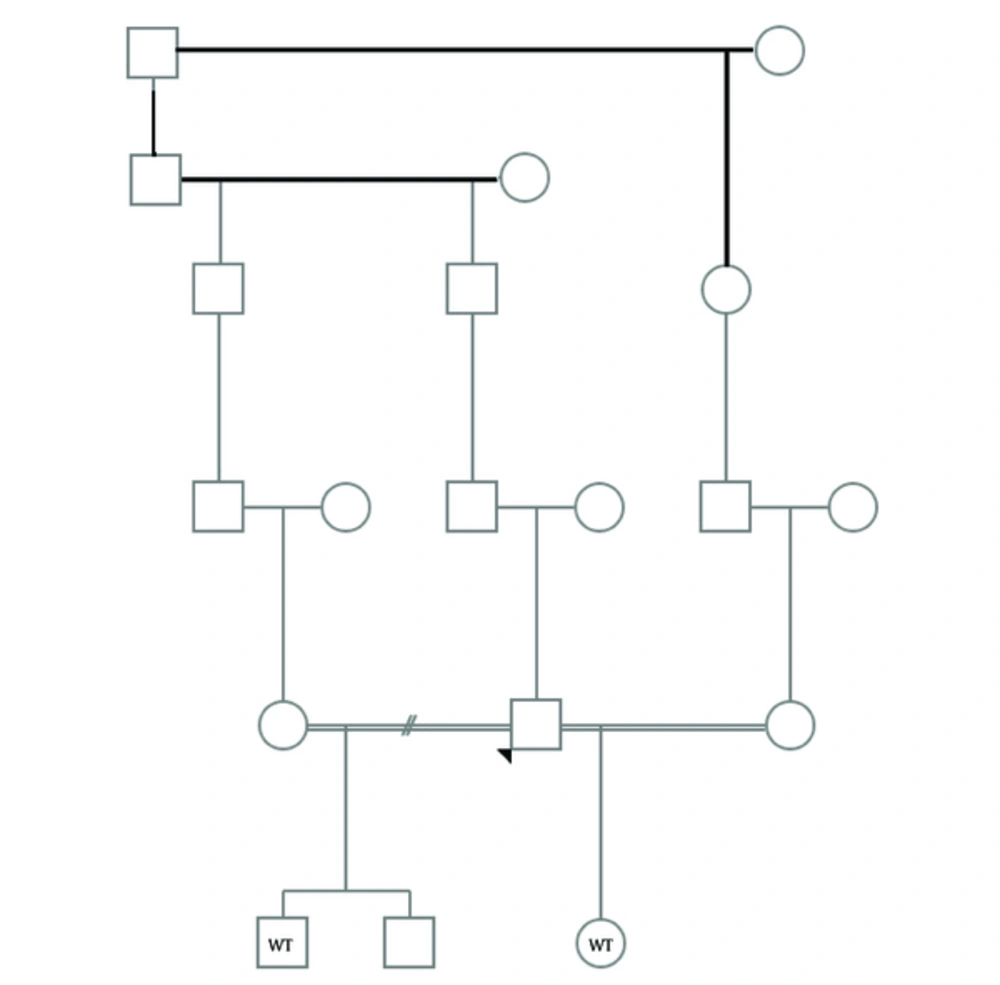
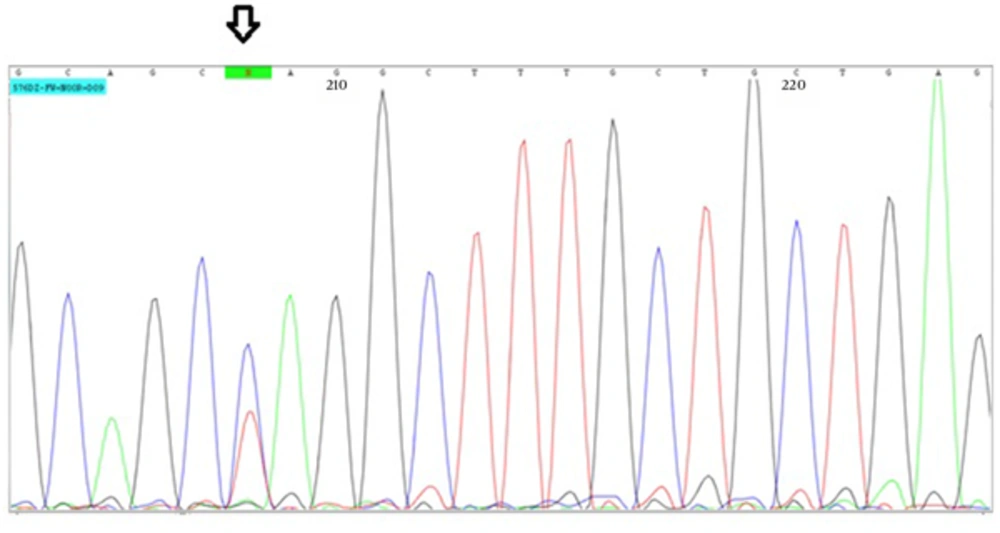
Here we report an Iranian family with two cases of WT. The proband has been referred to genetic counseling program because of a history of two affected children from two marriages with kidney tumor (Figure 1). The proband was phenotypically normal with no history of urogenital disorder. His son was 17 years old and got unilateral kidney cancer at the age of 3. His 1.5 year old daughter was recently diagnosed with bilateral kidney cancer and received appropriate chemotherapy. The results of the pathological reports of both patients showed the typical characteristics of Wilms’ tumor with blastemal, epithelial, and stromal components. No history of mental, growth retardation, or developmental delay has been reported in patients. In order to find the possible genetic background, bblood samples were collected from patients in EDTA tubes. Informed consents were obtained from parents before participation in the study in accordance with the protocol approved by local institutional ethics committee. DNA was isolated using the standard salting out method. Whole exome sequencing was performed using Illumina’s Genome Analyzer for the affected male patient with focus on 2752 OMIM disease genes (BGI-Clinical Laboratories, Shenzhen, China). A deleterious heterozygous stop gain mutation has been detected in WT1 gene (NM_000378, exon 3, c.C841T, p.Q281X). The results were verified by Sanger sequencing using the ABI Prism3130 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) in this patient (Figure 2). The presence of the detected mutation has been confirmed in the other patient by Sanger sequencing as well. The proband also showed the variant in heterozygote state. This variant has not been reported in generalist polymorphism databases (ExaC or exome variant server (EVS), dbSNP and 1000 genome project. Combined Annotation Dependent Depletion (CADD) tool which is a tool for scoring the deleteriousness of single nucleotide variants as well as insertion/deletions variants in the human genome showed that this variant would be deleterious with a score of 38.
Although this variant is a novel mutation, as it causes premature termination of the encoded protein, it is highly likely to contribute in the observed phenotype in the patients. The fact that the proband is heterozygote for the same mutation is in accordance with incomplete penetrance, which is commonly seen in tumor-causing mutations. This mutation is close to a similar truncating mutation in the exon 3 of WT1 gene that has been shown to cause WT (7).
A previous study has demonstrated that the penetrance rate for WT1 mutations is 100% for persons who received small WT1 mutations from their fathers, and is 67% for those who got small WT1 mutations from their mothers or large 11p13 deletions, or had de novo large 11p13 deletions regardless of the parental origin (1). In the present cases, there is no data regarding the parental origin of mutation in the proband. So although we demonstrated that both children of the proband have got the disorder, we cannot propose a mechanism for incomplete penetrance in the proband.
In brief, considering the variable penetrance of WT1 mutations depending on the parental origin (1), as well as the improved survival rates of patients with bilateral WTs (8), genetic counseling has a critical role in family planning for WT survivors and other family members.