1. Background
Colorectal cancer (CRC) is the third most common cause of cancer-related deaths in the world (1). The incidence of CRC is increasing in a number of Asian countries such as Iran during recent years (2, 3). Despite significant improvements in clinical therapy, most patients still die due to therapeutic resistance and recurrence of disease.
Cancer stem cells are unspecialized cells defined by two functional characteristics including stemness maintenance and multipotency. Some evidence indicates that “cancer stem cells” may be involved in tumor maintenance, therapeutic resistance, tumor progression, and distant metastasis. The CSCs have been identified in several solid tumors, including CRC (4). Despite their potential clinical significance, regulation of intrinsic CSC properties at the molecular level is poorly understood.
MicroRNAs (miRNAs) are a class of small noncoding RNAs that regulate gene expression by binding to 3’ un-translated regions of the complementary mRNA. This interaction leads to mRNA degradation or inhibition of protein synthesis (5). Previous studies have shown that miRNAs participate in several biological functions including development, cellular proliferation, differentiation and apoptosis. Moreover, new evidence has suggested that miRNAs could be involved in maintaining stemness of embryonic stem cells and CSCs. To date, several studies have described the role of miRNAs in cancer stem cell of lung, breast, hepatocellular, gastric, prostate, pancreatic, and Glioma (6). Experimental facts have suggested that some of the miRNAs including miR-126 (7), miR-181 (8), miR-372, miR-373 (9), miR-17-92 (10, 11), miR-21 and miR-205 (12) regulate the key properties of cancer stem cells including cell-cycle exit and differentiation, pro-survival and anti-stress mechanisms, epithelial-mesenchymal transitions (EMT) and migration and invasion.
Recent data suggest that miRNAs could implicate in sustaining stemness of colorectal CSCs; however, the role of miRNAs in human colorectal CSCs (CRCSCs) is still poorly understood. Investigation of miRNA expression profiles in colon CSCs and their potential role is crucial for elucidating characters of colon CSCs, which will help to develop anticancer drugs or novel therapeutic methods to target CSCs.
2. Objectives
The aim of this study was to investigate the miRNA expression profile in cancer stem cells compared to non-stem cells cancer cells
3. Methods
3.1. Tumor Cell Preparation
Colon cancer tissues were obtained from 3 patients (stage II, female, age range 58 - 65 years) after signing informed consent according to the ethics committee of Tarbiat Modares University. The histological diagnosis was based on microscopic features of carcinoma cells by expert pathologist that determines the histological types and grades of tumors. The studied patients did not receive any form of neoadjuvant therapy prior to surgery. Cancer tissues were intensively washed in PBS solution contains antibiotics and incubated overnight in DMEM/F12 (GIBCO) supplemented with penicillin (500 U/mL), streptomycin (500 mg/mL) (Invitrogen), and amphotericin B (1.25 mg/mL) (Sigma). Tumor tissue was mechanically and enzymatically dissociated after 2-hour incubation with Collagenase Type IV (1.5 mg/mL) (Invitrogen) at 37°C. Then, the single cells were used to be cultured in DMEM/F12 supplemented with 10% fetal bovine serum (FBS, GIBCO), to obtain primary colon cells, considered as parent cells and to be cultured in stem cell medium DMEM/F12 to obtain spheroids.
3.2. Cell Line and Culture Conditions
The human colonic adenocarcinoma cell line HT-29 was purchased from Iranian biological resource center (IBRC). Cells were cultured in DMEM/F-12 supplemented with 10% FBS and 1% penicillin/streptomycin in a humidified incubator at 37°C and 5% CO2. HT-29 Cells were cultured in completed media considered as parent cells. Cells were suspended using trypsinization (0.25% trypsin/EDTA (GIBCO) for 3 - 5 minutes).
3.3. Generation of Colonospheres
Single-cell suspensions from primary tumors and HT-29 cell line were cultured in ultra-low attachment 6 well plates in stem cell medium DMEM/F12 supplemented with 6 mg/mL Glucose, 5 mM HEPES, 4 mg/mL Heparin, 1%B27 (50X, invitrogen) , 1% (w/v) of non-essential amino acids (GIBCO), 1% (w/v) of L-glutamine (GIBCO), 10 ng/mL bFGF, 20 ng/mL EGF, 1% (w/v) of sodium pyruvate, 100 mg/mL apotransferrin, 25 mg/ml insulin, 9.6 mg/mL putrescin, 30 nM sodium selenite, and 20 nM progesterone (Sigma-Aldrich) 1% penicillin/streptomycin (GIBCO) to obtain spheroids. To evaluate the differentiation potential of stem cell-like cells, spheroids were cultured in DMEM/F12 supplemented with 10% FBS. All cell culture was carried out at 37°C in a 5% CO2 humidified incubator.
3.4. Clonogenic Assay
For colony formation assay, single-cell suspensions from colonospheres and parent cell-derived primary and HT-29 cells were seeded out at density of 500 cells/mL in a 12-well plate to form colonies for 5 - 7 days. Colonies were fixed with glutaraldehyde (6.0% v/v), stained with crystal violet (0.5% w/v) and colonies with over 50 cells were manually counted using a stereomicroscope.
3.5. Flowcytometry
Single cell suspension from colonospheres and parent cell-derived primary and HT-29 cells were stained according to protocol. Briefly, cells were detached by trypsin and re-suspended in PBS containing 1% BSA. After 10 minutes, cells were labeled with anti-human/mouse CD44-APC (eBioscience) and anti-human EPCAM-PE (Abcam). The cells stained with rat IgG 2b isotype control APC and mouse IgG 1 isotype control PE served as gating control. Flow-cytometry analysis was performed using FACS CantoII.
3.6. RNA Extraction and Quantitative Real-Time RT-PCR
Total RNA was extracted from the parent and colonosphere-derived primary, and HT-29 cells using high pure RNA isolation kit (Roche) according to the manufacturer’s instructions. Quantitative reverse transcriptase PCR was performed using light cycler RNA Master SYBR Green I (QIAGEN) on light cycler Roche version 3.5. The following genes were tested: c-myc, Klf4, Nanog, Sox2 and Oct4. The primer sequences and PCR conditions are summarized in Table 1. The relative amount of each mRNA was normalized to HGPRT. The fold-change from colonospheres relative to the control (parent) was calculated using 2-ΔΔCtmethod.
| Gene Name | Primer Sequences |
|---|---|
| c-myc | F: 5'-AGCGACTCTGAGGAGGAAC-3' |
| R:5'-CTGCGTAGTTGTGCTGATG-3' | |
| Sox-2 | F:5'-GGACTGAGAGAAAGAAGAGGAG-3' |
| R:5'-GAAAATCAGGCGAAGAATAAT-3' | |
| Oct-4 | F:5'-CGCCGTATGAGTTCTGTG-3' |
| R: 5'-GGTGATCCTCTTCTGCTTC-3' | |
| Nanog | F:5'-GCTAAGGACAACATT GATAGAAG-3' |
| R:5'-CTTCATCACCAATTCGTACTTG-3' | |
| Klf-4 | F: 5'-CCCAATTACCCATCCTTCC-3' |
| R:5'-GTGCCTGGTCAGTTCATC-3' | |
| HGPRT | F:5'-CCTGGCGTCGTGATTATGG-3' |
| R:5'-TCAGTCCTGTCCATAATTAGTCC-3' |
aCycle conditions were as follows: 1 cycle at 50°C for 10 minutes, followed by 1 cycle at 95°C for 10 minutes, followed by 40 cycles at 95°C for 10 seconds and 60°C for 1 minute.
3.7. In Vivo Xenografts Assay
To investigate whether tumor spheroid cells retain tumorigenic potential, single-cell suspensions from colonosphere and parent-derived primary and HT-29 cells were subcutaneously injected to 6 to 8 week-old nude mice. Cells were prepared at 1:1 ratio of PBS: collagen in a total volume of 100 µL, before injection. Mice were purchased from animal laboratory of Imam Khomeini Hospital and were maintained under standard conditions according to the guidelines of the animal care committee of cancer institute. In vivo studies were approved by the Animal ethics committee, Tarbiat Modares University. A portion of the tumor tissues was fixed in 10% formaldehyde and stained with H&E for histopathological examination.
3.8. MiRNA PCR Array
Total RNA from spheroid and parental cells-drived primary and cell line was extracted using Trizol (Invitrogen) and cDNA was synthesized by Universal cDNA Synthesis Kit II (Exiqon , Denmark ) according to the manufacturer’s instructions. MicroRNA expression profiling was performed using real-time PCR in 384-well format by the miRCURY LNA™ Universal RT MicroRNA PCR kit (Exiqon, Denmark) on Roche LC480. Expression levels of the microRNAs were measured using the comparative Ct method and normalized by miR-199a as endogenous control.
3.9. Statistical Analysis
Data are presented as means ± standard error of the mean (SEM). Statistical analysis was performed using SPSS 15 and GraphPad Prism 3.0. The paired t-test was used to analyze the median value of expression between the groups. Statistical significance was determined at P < 0.05. All tests were performed in triplicate.
4. Results
4.1. Generation and Validation of Spheroid Cells
According to previous experiments, we showed that single cells from primary tumors and HT-29 cell line formed spheroid colonies under serum free condition or stem cell media (13, 14) (Figure 1A). To investigate the capability of colon spheres differentiation, these cells were cultivated in 10% FBS medium. After one day, floating undifferentiated cells were attached to the plastic, gradually migrating from tumor spheres and differentiated into large and adherent cells.
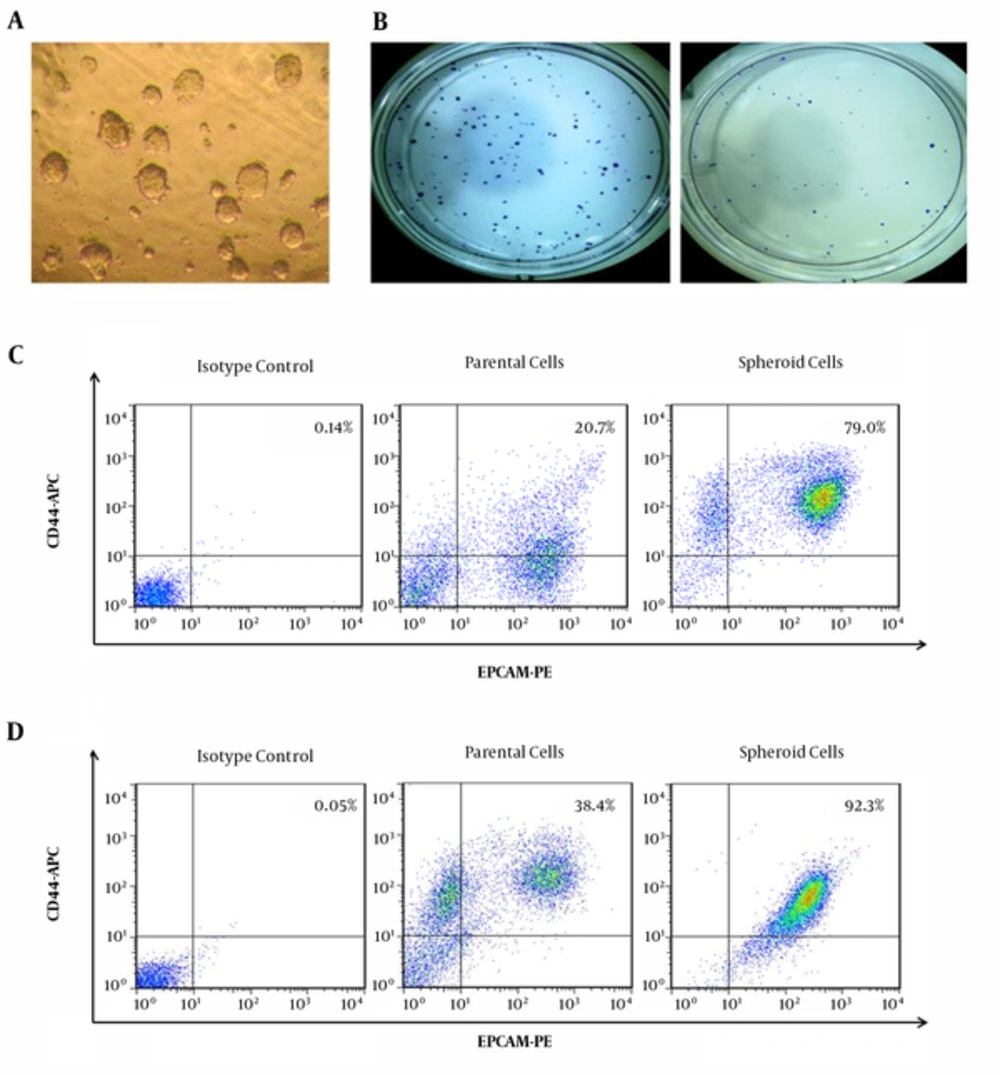
A, representative photographs show colon cancer cells expanded in vitro as undifferentiated spheres in serum-free medium containing EGF and FGF-2. Pictures show a typical colon sphere observed by the inverted phase contrast microscope in primary tumor (200x); B, analysis of cell colony numbers in clonogenicity assays of spheroid (left) and parent cells (right) in HT-29 cell line; C, spheroid populations formatted larger and more colonies compared with parent cells; D, representative scattered plots of flow cytometry analyses showing CD44 and EPCAM positive cells in parental and spheroid cells in primary tumors and in HT-29 cell line.
Spheroid cells expressed higher levels of EpCAM and CD44 molecules in comparison to parent cells. The results have demonstrated that 92% of tumor spheroid cells-derived HT-29 cell line were CD44+/EpCAM+, while matched parent cells only expressed 38% of CD44+/EpCAM+ markers. In case of the primary tumor, 79% of spheroid cells were positive for CD44/EPCAM, while 20% of parental cell showed CD44+/EPCAM+ (Figure 1C and 1D). These results suggested that colonospheres formed by the colon cancer cells have enriched population of CSCs.
To examine the clonogenic potential of the spheroid and parent cells from both primary and cell line, we plated and cultured the different cell population at clonal density for 7 days. The results of experiments indicated that spheroid cells possess a higher clonogenic activity than parent cells (P < 0.05). Our data showed that colony formation ability of spheroid cells from primary tumor was 185 ± 19 colonies/well vs. 76 ± 8 colonies/well parent cells (Figure 1B). Similar results were observed in colony formation ability of the spheroid derived HT-29 (196 ± 13 colonies/well vs. 58 ± 6 colonies/well) as compared with parent cells.
4.2. Spheroid Cells Exhibit Stemness Properties
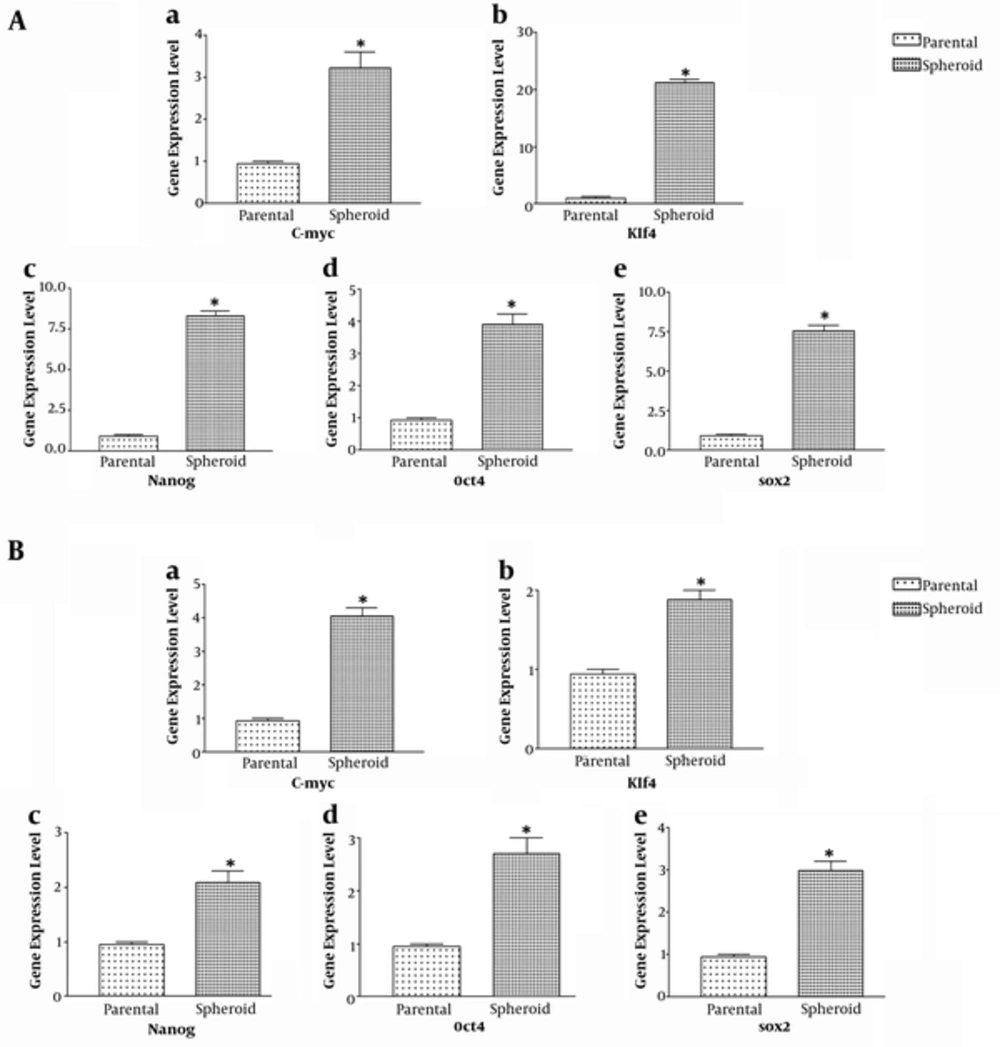
To evaluate the stemness properties, expression level of markers related to stem cell pathways including Klf4, Sox2, Nanog, C-myc and Oct4 were determined by RT-PCR in both spheroid and parent cells. As shown in Figure 2, spheroid cells derived primary and cell line expressed high levels of stemness genes as compared to parent cells (P < 0.05). Primary tumor derived spheroid cells also showed significantly increased level of these genes compared with the cell line derived spheroid cell (Figure 2).
4.3. Spheroid Cells Exhibit Tumorigenic Potential in Vivo
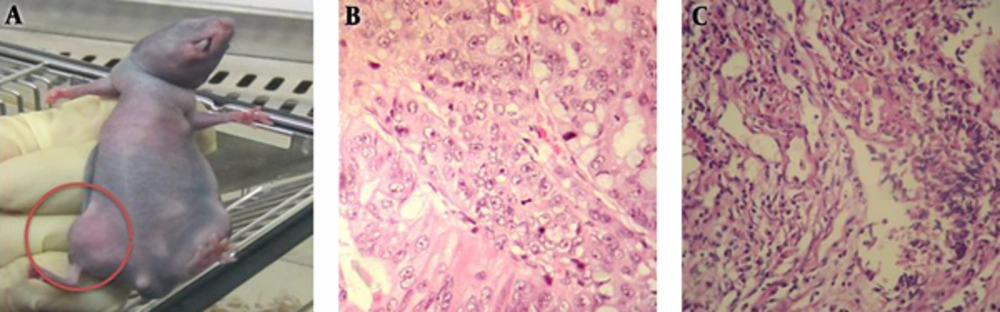
In line with previous studies, subcutaneous injection of low numbers of spheres confirmed the spheroid cells retain the capacity to initiate tumor growth in nude mice (13, 14) (Figure 3A). Our data showed that as few as 1000 spheroid cells from primary tumor were sufficient to obtain tumor growth; however, 1 ×106 parent cells were capable to form tumor in nude mice. Additionally, it was shown that at least 2,500 cells from spheroid cell derived HT-29 cell line were capable to form tumor (Table 2). The data, therefore, indicate that spheroid cells formed by the colon cancer cell are enriched in CSCs. Histopathological examination of xenografts that implicated from spheroid and parental cells showed that these tumors closely resemble the original human tumor (Figure 3B and 3C).
| Cell Type | Patients | Cell Number Injected | Tumor Incidence | Engraftment-Derived Spheres |
|---|---|---|---|---|
| Spheroid cells (HT-29) | - | 1 × 103 | 0/3 | - |
| 2.5 × 103 | 3/3 | Yes | ||
| 5 × 103 | 3/3 | Yes | ||
| Parental cells (HT-29) | - | 5 × 105 | 0/3 | - |
| 1 × 106 | 3/3 | Yes | ||
| Spheroid cells (primary tumor) | P1, P2, P3 | 1 × 103 | 1/3* | Yes |
| 2.5 × 103 | 3/3 | Yes | ||
| 5 × 103 | 3/3 | Yes | ||
| Parental cells (primary tumor) | P1, P2, P3 | 5 × 105 | 0/3 | - |
| 1 × 106 | 3/3 | Yes |
aP1 and P2 induced tumor in all of three experimented mice. But, P3 induced tumor just in one of the three mice.
4.4. Differential miRNA Expression Profile in Colon CSCs and Non-Stem Cells
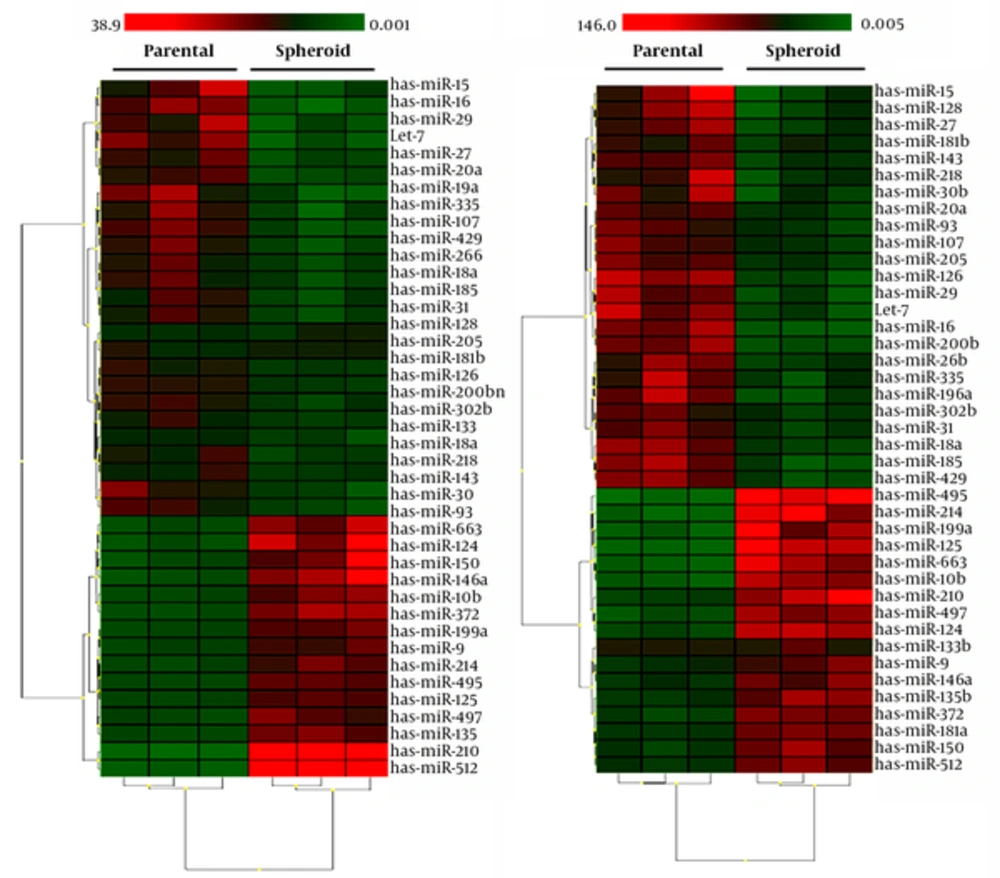
We used a PCR array to evaluate the differential miRNA expression profile in colon CSCs and non-stem cells. A total of 739 human miRNAs were examined. There are 39 differentially expressed miRNAs, 24 of them had lower mean expression in the cancer stem cells samples. By contrast, 15 miRNAs had higher expression levels in CSCs samples (Figure 4). Of these, miR-495, miR-125, miR-199a were the most significantly up-regulated miRNA. Interestingly, miRNA expression profile in primary cancer stem cells compared to HT-29 derived cancer stem cells was different. Expression of some miRNAs including miR-125, miR-124, miR- 495, miR- 497, miR-663, miR-214, miR-10b, and miR-199a significantly increased in comparison to primary tumor derived CSCs. By contrast, down-regulation of miR-128, miR-126, miR-205, and miR-200b without noticeable changes in primary tumor derived CSCs were observed.
5. Discussion
In contrast to traditional models of cancer biology, the cancer stem cell model proposes that only a few fractions of cells within a tumor microenvironment have the potential to initiate and sustain tumor growth known as cancer stem cells (CSCs) (15). Genetics and epigenetic control as well as signaling from neighboring cells regulate stem cells properties. MiRNA levels control self-renewal, pluripotency and differentiation properties of stem cells. Enhancement of epigenetic abnormalities due to failure in repair errors leads to some signaling cascades that support tumorigenesis and creation of cancer stem cells (16). Isolated cancer stem cells from primary and HT-29 cancer cells express important factors in stemness-keeping and decisive properties of progenitor cells including Sox-2, Oct-4 and Nanog that were previously reported to be involved in stemness maintenance in human CSCs (17).
Various stages of colon cancer have showed different miRNA expression profiles. These data show that miRNAs regulate special phenotype and biological properties of CSCs (18). So, miRNAs profiling in colon CSCs in comprasion to non-stem cells help to clarify miRNAs roles in progression of cancer.
We isolated CSCs according to previous studies, enriched CSCs with spheres creation and documented using CD44+ and EPCAM+. Here, we profiled primary and HT-29 cancer stem cells compared to parent colorectal cancer samples and found 39 differentially expressed miRNAs, twenty four of which had lower mean expression in the cancer stem cells samples. By contrast, 15 miRNAs had higher expression levels in CSCs samples suggesting their function to block tumor suppressor genes or to activate oncogenes in colorectal cancer. One of the over expressed miRNAs in CSCs was 181a that correlates with induction of TGF-β epithelial to mesenchymal transition (EMT) through smad-7 transcription factor (19). Similarly, miR-302 was down-regulated in CSCs that block TGF-β mediated EMT (20). Another miRNA which promotes EMT is miR-125b that was up-regulated in CSCs showing that TGF-β EMT pathway is commonly activated in CSCs and plays important roles in CSCs properties. MiR-125b also inhibits apoptosis by affecting p53 and its target genes p21 and Puma (21). Another up-regulated miRNA that is involved in apoptosis inhibition is miR-372 through down-regulation of a tumor suppressor gene, LATS-2 (22).
CSCs indicate some special characteristics such as continuous cell proliferation and ability to colony formation which are regulated by miRNAs. In the current study miR-495, miR-214 and miR199a up-regulated in CSCs and were involved in cell proliferation that was previously reported in breast and ovarian cancers (23-25). MiR-495 promotes cell proliferation and oncogenesis via down-regulation of E-cadherin and REDD-1 (23) but miR-214 affects the p53-Nanog axis (24). Furthermore, enhanced proliferation and maintenance of HT-29 derived CSCs compared to primary tumor derived CSCs could be related to miRNAs including miR-495, miR-214 and miR-199a.
Cancer stem cells involve in invasion and metastasis through different genes regulated by miRNAs. MiR-9, miR-10b and miR-663 have critical roles in invasion and migration. MiR-9 has a dual function and plays a critical role in local recurrence of cancer (26). By contrast, some studies showed that it decreases cell viability and increases apoptotic activity through blocking anti-apoptotic gene, MTHFD-2 (27). MiR-10b targets KLF4 leading to increased migration and invasion as well as enhancing pro-metastatic gene products including RhoC, urokinase plasminogen activator receptor (uPAR), α3-integrin, and MT1-MMP (28). One of the other important up-regulated miRNAs in CSCs derived from both primary and HT-29 cancer cells was miR-146a. Previous studies indicated that miR-146a may play important roles in cell survival and multi drug resistance (29).
In this study, miR-429, miR-196, miR-218, miR-18a, and miR-26b as well as miR-15 and miR-16 were decreased in CSCs. MiR-429 modulate migration through ets-1 (30). On the other hand, miR-218 targets oncogene, Bmi-1 (31). Therefore, down-regulation of miR-429 and miR-218 increase stem cells proliferation and migration. Also, a previous study showed mir-18a acts as tumor suppressor by targeting K-ras which plays a critical role in cancer cells’ growth and cell proliferation (32). One of the most important down-regulated miRNAs is miR-26b that causes the significant suppression of the cell growth and the induction of apoptosis in cancer cells (33). Other important miRNAs including miR-15, miR-16, miR-181b and miR-196a induce apoptosis by targeting bcl-2 and their down-regulation led to maintenance of cancer stem cells (34-36).
In summary, we showed that different miRNA expression in both primary and HT-29 colon CSCs indicate stemness maintenance of colon CSCs may be largely regulated by miRNAs. This study adds further evidence to prove miRNAs’ roles in colon cancer invasion and recurrence and the findings of the current study may contribute to the treatment of colon cancer.