1. Background
Cancer is characterized as a group of diseases induced by uncontrolled cell growth and stimulated tumor metastasis and formation (1). Worldwide, lung cancer is the leading cause of death associated to cancer. Historically, lung cancer is divided into two important categories, non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC), with the former consisting mostly of squamous cell carcinoma, adenocarcinoma, and large cell carcinoma (1). Generally, 75% to 85% of global lung cancer cases are NSCLCs. At present, more than 70% of people with NSCLC have highly advanced cancer (2, 3). Lung carcinogenesis occurs from a variety of factors, such as deactivation of tumor suppressor genes or activation of the oncogenes. As a result, the molecular differentiation of NSCLC and SCLC is uncertain. So, understanding these mechanisms will be developing therapeutic approaches for the treatment of human NSCLC (4-6).
MicroRNAs (miRNAs) are small single-stranded RNA molecules with 18 to 25 nucleotides in length. They have the potential to modulate gene expression at the post-transcription level. miRNA can inhibit and degrade target gene expressions by binding to the 3′ untranslated region and produce the complex on the targeted gene. In the past decade, the regulatory role of miRNAs has been studied on cancer biology. In normal cells, miRNAs control genes that are involved in cellular growth, differentiation, proliferation, and apoptosis. Unsurprisingly, miRNAs, which prevent genes from involving in cell cycle progression, are often down regulated in cancer cells (7-9). Different studies indicate that miRNAs are potential molecules for cancer therapy, because they have tumor suppressor functions and potential biomarkers for cancer diagnosis (10, 11).
miRNA-21 (miR-21), one of the first recognized and best-established miRNAs in human cells, has been studied in various diseases such as cancer and cardiovascular diseases. Evidence has demonstrated that in lung cancer, the expression of miR-21 is controlled by epidermal growth factor receptor signaling (12-14). miRNA-205 (miR-205) is found in a lung cancer-related genomic amplification at 1q32.2 region. Recently, it has been revealed that the loss of miR-205 stimulates the transition of epithelial tissue to mesenchymal tissue during tumor progression. A previous study has suggested that SMAD4 expression is regulated by miR-205 through targeting its 3’-UTR in NSCLC (15).
2. Objectives
Our study investigated the expression of miR-205 and miR-21 in NSCLC and compared it to healthy controls.
3. Methods
3.1. Sample Collection
Fifty blood specimens of patients with lung cancer, who received surgery resection or biopsy for NSCLC at the Department of Clinical Laboratory, Masih Daneshvari Hospital (Shahid Beheshti University of Medical Sciences, Tehran, Iran) and 50 blood sample of healthy individuals were collected.
The healthy sample featured normal bronchoscopy reports-no history of illness, no smoking, and no history of using anticancer drugs. Cancer sample patients were those in the early stages with no distant metastasis. Chemotherapy, radiotherapy, and surgery were not performed on any patients.
All samples were finally confirmed with histopathology examinations. All the protocols used in the present investigation, including human subjects, were initially reviewed and approved by the Ethical Review Board of Masih Daneshvari Hospital. Additionally, in accordance with the Declaration of Helsinki, written informed consent was obtained from each participant. The clinical pathology characterizations are below in Table 1.
| Variable | Number of Patients | miR-21 Expression | P Value | miR-205 Expression | P Value |
|---|---|---|---|---|---|
| Age | 0.751 | ||||
| ≤ 35 | 8 | 4.12 | 6.36 | ||
| 35 - 50 | 10 | 4.78 | 0.790 | 6.22 | |
| 51 - 65 | 18 | 4.56 | 6.31 | ||
| > 66 | 14 | 4.25 | 6.12 | ||
| Gender | 0.548 | ||||
| Male | 24 | 4.35 | 0.844 | 6.45 | |
| Female | 26 | 4.23 | 6.31 | ||
| Stage | 0.892 | ||||
| I | 9 | 4.13 | 6.61 | ||
| II | 12 | 4.23 | 0.812 | 6.38 | |
| III | 11 | 4.65 | 6.47 | ||
| IV | 18 | 4.48 | 6.71 | ||
| Lymphatic metastasis | 0.643 | ||||
| Negative | 27 | 4.56 | 0.678 | 6.71 | |
| Positive | 23 | 4.35 | 6.44 | ||
| Differentiation | 0.89 | ||||
| Degree | 12 | 4.33 | 6.51 | ||
| Low | 17 | 4.37 | 0.99 | 6.42 | |
| Middle | 21 | 4.81 | 6.22 | ||
| High | 6.51 | ||||
| Surgery | 0.521 | ||||
| Yes | 24 | 4.55 | 0.551 | 6.71 | |
| No | 26 | 4.12 | 6.38 |
3.2. PBMC Isolation
For prepared total peripheral blood mononuclear cells (PBMC) isolation, 5 mL blood samples were mixed with anticoagulant. PBMCs were isolated from whole blood samples by using Ficoll-Paque Plus (GE Healthcare, United Kingdom) density gradient centrifugation, based on the instructions provided by the manufacturer. In brief, whole blood samples were layered on Ficoll-Paque Plus solution and centrifuged at 800 × g at 4°C for a duration of 30 minutes. Then, PBMC layer was extracted and washed twice with phosphate buffered saline (PBS) and centrifuged at 350 × g at 4°C for 10 minutes. After washing, PBMCs were re-suspended in PBS, aliquots stored at -80°C until analysis.
3.3. Total RNA Extraction and cDNA Synthesis
Total RNA was extracted from PBMCs, using Trizol reagent (Invitrogen, Life Technologies, Germany), according to manufacturer’s instructions. The first-strand complementary DNA (cDNA) was synthesized, using a kit procured from Takara, Japan (PrimeScriptRT Reagent Kit), as per the manufacturer’s recommendations.
3.4. miRNA Quantification by Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
miRNAs were quantified via qRT-PCR, using U6 miRNA as control. The qRT-PCR was carried out, using a SYBR Premix ExTaq Kit (Takara) on a QiagenRotor-Gene (Qiagen, Germany).
The real-time RT-PCR reaction components included (A) 2 µL of the template, (B) 4 µL of the Master mix, (C) Primer with optimal concentration found in set up tests, and (D) Deionized distilled water to reach a final reaction volume of 20 µL.
Briefly, the data were obtained as the threshold cycle (Ct) value, and the ΔCt values were determined by subtracting the average GAPDH Ct value from the average target gene Ct value. Relative expression of each gene was calculated by 2-ΔΔCt formula.
3.5. Statistical Analysis
Statistical tests were carried out via the Mann-Whitney test, adopted for comparing miRNAs expression difference between NSCLC patients and healthy controls. All analyses were performed with SPSS 21.0 software, and probability P < 0.05 was considered to get the statistically significant results.
4. Results
4.1. PBMC miR-205 and miR-21 Expression Levels in Lung Cancer in Comparison to Healthy Controls
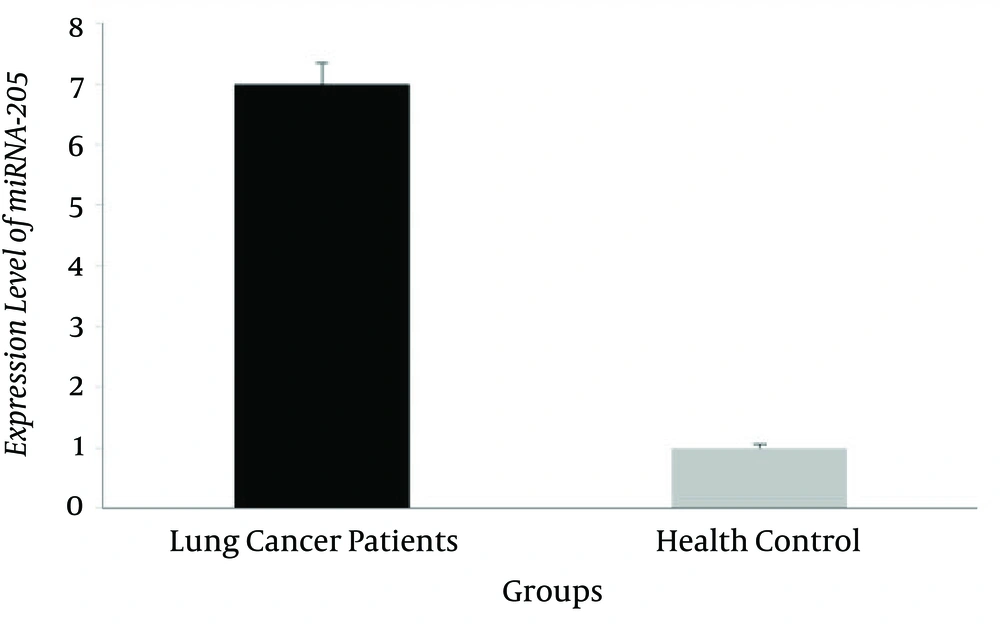
The miRNA-205 level of expression in 50 pairs of specimens, related to patients with tumor and healthy blood samples, were assessed by quantitative real-time PCR. Our results revealed that the miR-205 level of expression in the patients with lung cancer significantly increased in comparison to healthy controls (6.8 ± 0.42 and 1.2 ± 0.19, respectively), (P = 0.014; Figure 1).
The expression of miR-21 in lung cancer (5.2 ± 0.52) compared to the control group (1.6 ± 0.14) significantly increased (P < 0.001; Figure 2).
4.2. miR-21 and miR-205 Expression Level in Lung Cancer Samples in Comparison to Clinicopathological Features
The analyzed relationship of miR-21 and miR-205 with lung cancer clinicopathological features included clinical stage, age, gender, lymphatic metastasis, differentiation degree, and surgical history. There is no statistical correlation to both miR-21 and miR-205 levels in PBMCs with clinical pathology (Table 1).
5. Discussion
Although there has been an incremental development in the survival rate of NSCLC patients, after decades of investigation, there is still a lack of real biomarkers for lung cancer diagnosis. For most cancers, blood-based proteins have been confirmed and used as biomarkers in clinical diagnosis (7). Therefore, the discovery of lung cancer diagnostic biomarkers is a critical imperative. miRNAs, a cluster of regulatory small molecular RNAs, have been associated with the progression of several cancers and some of them have been described as a potential biomarkers in cancer diagnosis.
Previously, investigations have reported that the abnormal expressions of miRNAs in NSCLC tissues depend on the normal tissue of lungs (8). Initially, miR-21 was renowned as one of the most important apoptotic suppressors of miRNA in different cell linings (16). Later, a large-scale study, conducted in 540 human samples, introduced miR-21 as the only miRNA overexpressed in stomach, lung, prostate, colon, breast, and pancreas cancers. Following this report, another study recognized miR-21 as an oncogenic miRNA and demonstrated its overexpression in most types of cancer (14, 17). However, others have indicated that miRNAs in patient blood samples are either related to proteins such as lipoproteins and Argonaut (18-20) or enclosed within cellular fragments elected as exosomes, macrovesicles, micro particles, or extracellular vesicles (21, 22). Moreover, from 2008 to 2013, 154 circulating extracellular miRNAs were detected in 26 different tumor types. In this study, our research discovered that miR-21 detected in PBMCs was significantly elevated in patients with lung cancer and could be a potential biomarker for lung cancer diagnosis. Our research study also found that miR-21 is applicable for NSCLC in all stages as a biomarker. Nevertheless, there are no parallel results between the PBMC miRNA profile of NSCLC and the circulating miRNAs in extracellular plasma or serum of patients with lung cancer (23, 24). Therefore, miRNA expression in PBMCs is not similarly affected by circulating tumor cells. In addition, PBMC expression of miR-21 has been shown to be associated with other cancer types. For instance, a few studies have suggested that the assessment of miR-21 level in PBMCs could highlight cancers with the role of an anti-apoptotic factor (16). Besides, there is no similarity of the differentially expressed miRNAs of PBMC between NSCLC and other types of tumors.
Our findings revealed that the expression levels of miR-205 in the patients with lung cancer increased in comparison to healthy controls. Verdoodt et al. have found that miR-205 is down regulated in prostate cancer and can be a novel regulator of the anti-apoptotic protein Bcl2 (25). Lung cancer is one of the earliest cancers that needs new biomarkers for its early diagnosis and has a poor prognosis and a mortality rate of thousands of patients per year (2).
Taken together, significant progress has been made in promoting the signatures of miRNA into new biomarkers for different types of cancers, especially lung cancer. It seems that miRNA signatures are efficient markers for early diagnosis or anti-cancer therapy. However, a major challenge for cancer detection is the implementation of standardized protocols for the analysis of miRNAs. In addition, cancer therapy needs specific delivery systems for transporting miRNAs to the tumor area. Overall, a better insight into the biological function of miRNAs in cancer cells contributes not only to cancer detection, but also to cancer therapy.
5.1. Conclusions
Overall, significant progress has been made in promoting the signatures of miRNA into new biomarkers for different types of cancers, especially lung cancer. It seems that miRNA signatures are efficient markers for early diagnosis or anti-cancer therapy. However, a major challenge for cancer detection is the implementation of standardized protocols for analysis of miRNAs. In addition, cancer therapy needs specific delivery systems for transporting miRNAs to the tumor area. Overall, a better insight into the biological function of miRNAs in cancer cells contributes not only to cancer detection, but also to cancer therapy. So, the present study is a basic study on the expression of MiRNA-21 and miRNA-205 in NSCLC patients. Large-scale and long-term follow-up studies are encouraged to confirm the significance of this potential biomarker in NSCLC.