1. Background
Acute lymphoblastic leukemia (ALL) is the most frequently encountered cancer and blood malignancy among children. ALL is caused by abnormal development and uncontrolled proliferation of lymphoid progenitor cells or hematopoietic stem cells and may be of a B-lineage (B-ALL) or a T-lineage (T-ALL) in the bone marrow (1). T cell acute lymphoblastic leukemia (T-ALL). Malignant transformation of T-cell precursors results in T-cell ALL (T-ALL), occurring in almost 25% of adults and 15% of children (2).
T-ALL is normally associated with a large tumor showing hyperleukocytosis. In addition, It is associated with mediastinal enlargement and risk of central nervous system involvement (3). Although many treatments have been discovered for T-ALL, it is associated with recurrence in many cases. Furthermore, the development of resistance to chemotherapy drugs is another problem. Therefore, it is necessary to find a new treatment, which can be used besides chemotherapy and radiotherapy to control T-ALL tumors (4).
There are numerous side-effects often related to chemotherapy drugs; therefore, RNA interference (RNAi) can reduce these effects because of its specificity and potency. In this method, appropriate personalized drugs can be developed for a patient so as to increase its effectiveness (5).
RNAi or mechanisms regulating gene expression at the post-transcriptional level are mediated by small noncoding RNA, such as small interfering RNA (siRNA), antisense oligonucleotides (ASOs), piwi-interacting RNA (piRNAs), and microRNA (miRNA) (length, 18 - 30 nucleotides) (6). Generally, siRNA is originated from double-strand RNA (dsRNA) with homologous complementary sequences of specific mRNA. The antisense strand attaches to the target mRNA, while double-stranded siRNA unwinds; the specific mRNA is degraded leading to phenotype dysfunction (7). Recently, this technology has been used in gene therapy and gene function research (8). Therefore, oncogenes, mutations in tumor suppressor genes, and besides other genes are involved in the progression of tumors and can be suggested as proper targets for RNAi gene silencing (5).
Deficiencies in the apoptotic pathway is a hallmark of cancer cells (9). Myeloid cell leukemia-1 (Mcl-1) from the Bcl2 family is a pro-survival protein. The Mcl-1 gene, with 3 exons, is located on chromosome 1q21. In 1993, Kozopas et al. discovered Mcl1 gene during differentiation of ML-1 into monocyte and macrophage. Mcl-1 is majorly involved in cell differentiation, cell death, and cell cycle program (10). This protein is overexpressed in different lymphoid and hematopoietic cancers, as well as solid tumors including CNS, lung, ovarian, colon, breast, melanoma, prostate, and renal tumors (11, 12). Based on the recent reports Mcl-1 is necessary for the survival of hematopoietic and tumor cells (13). Mcl-1 has been recognized as an important factor in common cancer therapies. It can be a potential target in many cancers since its downregulation is adequate to promote cell death in cancer cells (12).
2. Objectives
In the present study, siRNA Mcl-1 was transfected into the Jurkat cell line, which is expressing high levels of Mcl-1, to evaluate the effects of Mcl-1 gene suppression on the survival and cell death in ALL. Apoptosis was induced by silencing Mcl-1 expression in the Jurkat cell line.
3. Methods
3.1. Cell Culture
In this study, Jurkat cell line, which purchased from Pasteur Institute of Tehran (Iran), was kept in RPMI-1640 medium (Sigma-Aldrich, USA), containing 1% antibiotics (including 100 IU/mL of penicillin and 100 μg/mL of streptomycin; Sigma-Aldrich), 10% heat-inactivated fetal bovine serum (FBS; Sigma-Aldrich), 1% sodium pyruvate, and 2 mM glutamine in a humidified 5% CO2 atmosphere at 37°C. At an initial concentration of 1×105 cells/mL, the cells were subcultured when reaching 80% - 90% confluence and then applied in the logarithmic growth phase.
3.2. siRNA Transfection
The negative control (NC) siRNA and Mcl-1 siRNA were supplied by Santa Cruz Biotechnology. The Jurkat cells were cultured in the RPMI-1640 medium (without FBS and antibiotics) in 6-well plates right before transfection at a density of 2×106 cells/well. Based on the instructions of the manufacturer, siRNA was used for transfection. In brief, after dilution of the transfection reagent and siRNA in the siRNA transfection medium (Santa Cruz Biotechnology) separately, they were gently mixed. After combining the solutions, they were incubated for 30 minutes to form a complex at ambient temperature.
The complex liquid, containing the reagent and siRNA, was gradually added to each well (containing medium and cells confluence, 80% - 90%) with 800 µL of the optimal medium and incubated in a chamber in a humidified CO2 atmosphere for 6 hours at 37°C. After adding RPMI-1640 medium (1 mL) containing 20% FBS, without removing the transfection mixture, the cells were incubated under the aforementioned conditions. After 48 hours, the cells were collected for further evaluation.
3.3. RNA Isolation and Q Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Based on the instructions, a total RNA purification mini kit (Yekta Tajhiz Azma, Tehran, Iran) was employed for extracting total RNA from Jurkat cells. RNA (1 µg) was reverse-transcribed into cDNA with 1 µL of Moloney murine leukemia virus reverse transcriptase (M-MLV RT), 1 µL of dNTPs (10 mM each), 4 µL of 5X reaction buffer, 1 µL of random hexamer primer, and 0.6 µL of RNasin (40 U/µL). Afterward, qRT-PCR was performed to amplify 1 µL of cDNA, using SYBR Green-1 dye on an ABI 7300 system (Applied Biosystems, NJ, USA) and a sequence detection system (Corbett Life Science, NSW, Australia). Based on the fluorescent signal the binding of SYBR Green dye to ds DNA reflects the generated amount of ds DNA product during PCR. SYBR Green I dye was used to enhance sensitivity and specificity. PCR specificity was confirmed, using melting curve analysis.
The PCR assay was carried out using 5 µL of SYBR green reagent, 0.2 µL of passive reference dye (50X), 1 µL of cDNA template, 0.5 µL of each primer, and 2.8 µL of nuclease-free distilled water; the reaction volume was 10 µL. In this study, the primers included: Mcl-1, 5′-GCGACGGCGTAACAAACTG-3′ (forward) and 5′-GAACTCCACAAACCCATCCCAG-3′ (reverse) with an amplified fragment length of 191 bp; and β-actin, 5′-TCCCTGGAGAAGAGCTACG-3′(forward) and 5′-GTAGTTTCGTGGATGCCACA-3′(reverse) with an amplified fragment length of 131 bp. The conditions of cDNA amplification using PCR were as follows: pre-degeneration for 3 minutes at 95°C, degeneration for 30 seconds at 95°C, annealing for 25 seconds at 64°C, extension for 30 seconds at 72°C (40 cycles), and storage at 4°C. Using the 2 -(ΔΔCt) method, relative Mcl-1 mRNA expression was determined, with β-actin mRNA expression as the internal control. The PCR reactions were performed in triplicate.
3.4. Cell Cytotoxicity Assay Using MTT
The MTT assay kit (Sigma-Aldrich; USA) was used to determine the cytotoxic effect of Mcl-1 siRNA on the Jurkat cell line. Cells in the logarithmic growth phase were collected and counted. Afterward, the cells (4×104 cells/well) were cultured in 96-well plates. Subsequently, transfection was performed according to the above-mentioned method. The groups included different doses of Mcl-1 siRNA (40 - 80 pmol), untreated cells, cells treated with scrambled siRNA (negative control), pure Mcl-1 siRNA, and transfection reagent. The MTT solution (5 mg/mL) was added at 48 hours post- transfection (volume, 20 µL/well). In the process of incubation, water-insoluble formazan crystals were formed within 4 hours at 37°C. Then, 100 µL of the supernatant was carefully removed from the wells, and DMSO (100 µL; 0.5 mg/mL) was added to solubilized crystals. Optical density (OD) was read by a microplate reader (Rayto, China) (Awareness Technology, FL, USA) in each well at 570 nm (reference, 650 nm).
3.5. Data Analysis
Data are presented as mean ± SEM. Data analysis was performed utilizing GraphPad Prism-5 statistic software (LaJolla, CA, USA) and one-way analysis of variance (ANOVA) was used to determine variations among groups. P < 0.05 was statistically accepted to be significant.
4. Results
4.1. Mcl-1 Expression in Jurkat Cell Line Following siRNA Treatment
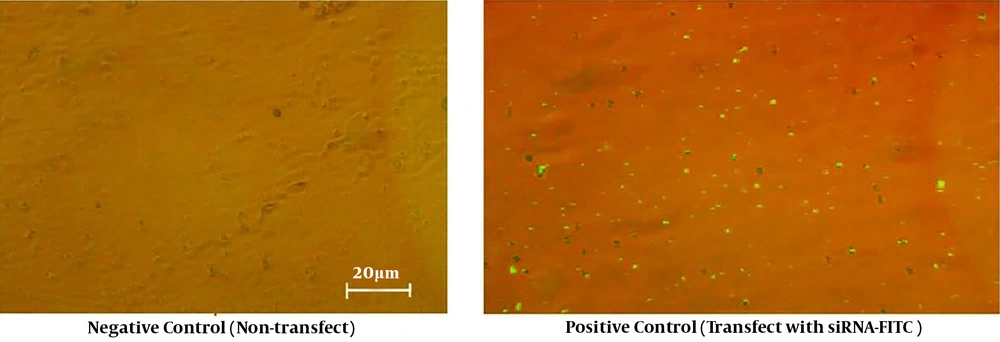
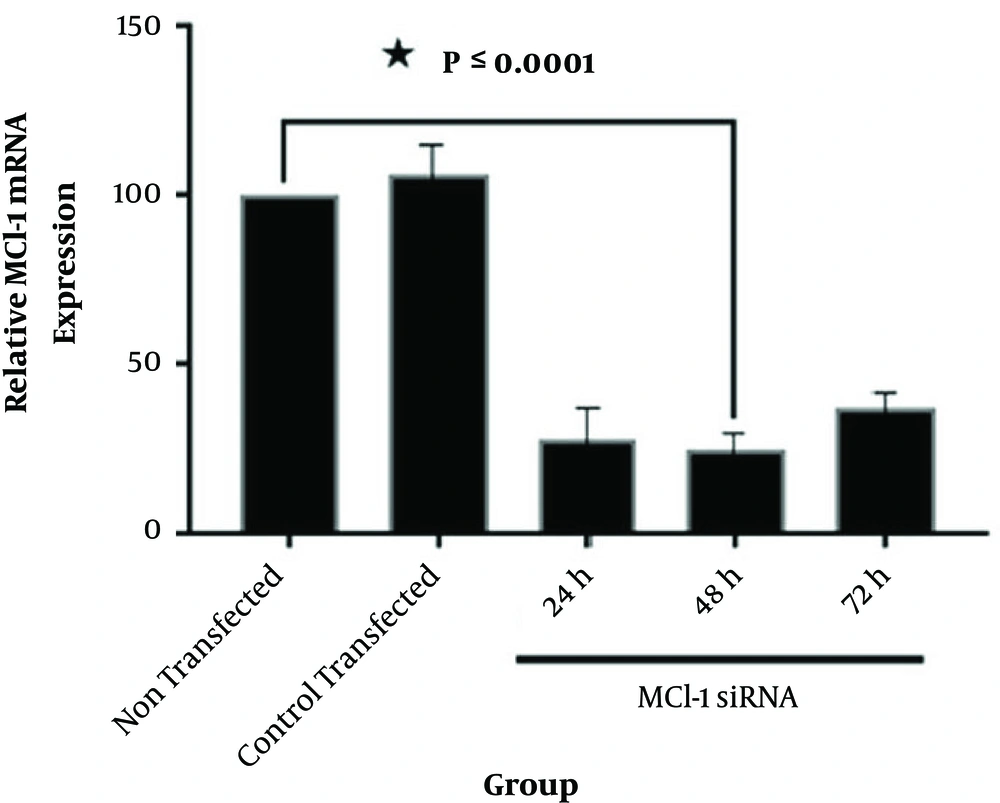
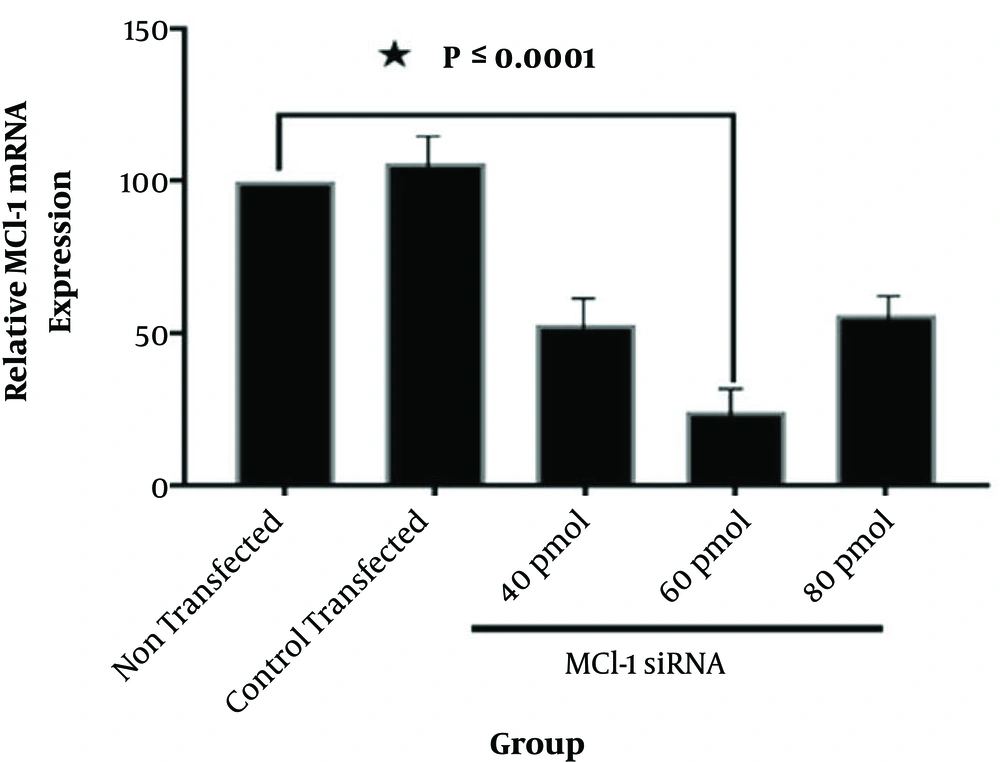
The control siRNA, tagged with fluorescein (siRNA-FITC), was used to assess transfection efficiency (Figure 1). According to Figures 1 and 2, Mcl-1 siRNA reduced Mcl-1 mRNA both dose- and time-dependently (P < 0.0001). The relative Mcl-1 expression was 27.8%, 24.7%, and 37.1% after 24, 48, and 72 hours of transfection, respectively (P < 0.0001; Figure 2), while the relative expression of 40, 60, and 80 pmol of Mcl-1 siRNA on Mcl-1 mRNA was 52.9%, 24.3% and 56.2%, respectively (P < 0.0001; Figure 3). The optimum knockdown time and concentration were considered respectively as 48 hours and 60 pmol. In addition, the level of β-actin mRNA expression (internal control) was similar in qRT-PCR for all the groups. Transfection with NC siRNA (scrambled control, a sequence without specific degradation of cellular mRNAs) insignificantly affected Mcl-1 expression in comparison with the control group. According to the results, specific siRNA could target Mcl-1 mRNA and reduce Mcl-1 gene expression in Jurkat cells.
The Jurkat tumor cells transfected with Mcl-1 siRNA at different times. Total RNA was extracted and mRNA was analyzed via qRT-PCR assay at 24, 48, and 72 hours following transfection. The relative mRNA expression was quantified by 2-(ΔΔCT) formula (β-actin, internal control) (data presented as mean ± SD; * P < 0.0001 vs. controls).
The Jurkat tumor cells transfected with different doses of siRNA (40, 60, and 80 pmol). Total RNA was extracted and mRNA was analyzed via qRT-PCR at 48 hours after transfection. The relative expression of mRNA was determined with 2-(ΔΔCT) formula (β-actin, internal control) (data presented as mean ± SD; * P < 0.0001 vs. controls).
4.2. Dose-Dependent Cytotoxic Effects on Jurkat Cells Induced by Mcl-1 Suppression
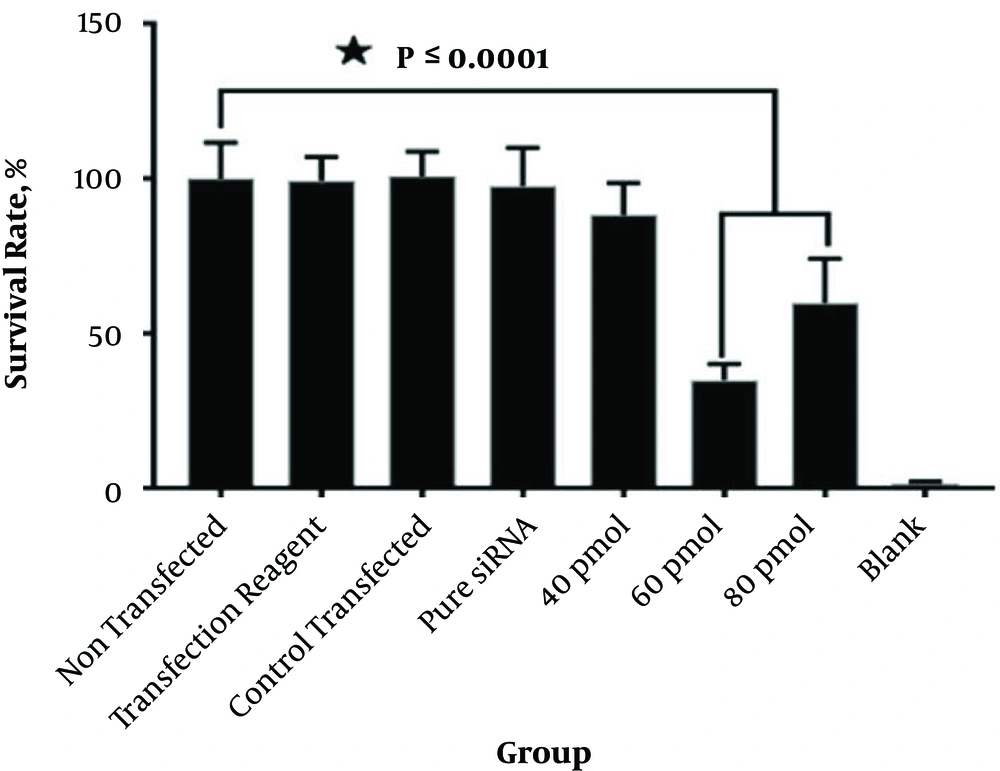
According to Figure 3, the percentages of survival were 88.33%, 35.05%, and 59.88%, respectively. Cytotoxicity significantly improved with 60 and 80 pmol of Mcl-1 siRNA-transfected cells versus the blank control and scrambled siRNA-transfected cells (P < 0.0001). In contrast, compared with the control and non-transfected groups, cell survival slightly reduced with 40 pmol of Mcl-1 siRNA. Moreover, transfection with control transfected siRNA and pure siRNA and transfection reagent had no significant cytotoxic effects on leukemic cells in comparison with the blank control group (Figure 4).
5. Discussion
T-ALL is a malignant hyperplastic disease of the hematopoietic system and it is the most common tumor in children and adolescents (14). Considering the development of chemoresistance in leukemia cells, most patients, exposed to standard treatments, show relapse following the initial treatment or fail to achieve complete remission (15-17). Improvement of treatment methods has caused a significant increase in survival; nevertheless, the survival of adult T-ALL is very low (< 40% for > 60 years). These studies show that non-toxic medicines and new strategies are required to improve therapy and increase survival in T-ALL patients (14). Gene therapy is an effective molecular targeted therapy. In recent years, RNAi technology has been the most common and simple genetic tool (18).
For instance, siRNA, which disrupts the target gene expression, is an overexpressed specific gene in tumor cells. It can inhibit tumor formation, growth, and proliferation (19-22).
Overexpression of some oncogenes regulates cellular proliferation and inhibits apoptosis in different cancers, such as leukemia and breast cancer (23-26).
Apoptosis is controlled by 2 signaling pathways. The extrinsic pathway, which is activated by extracellular death receptors, causes caspase-8 activation, while the intrinsic mitochondrial pathway is stimulated by many signals, leading to mitochondrial outer membrane permeabilization (MOMP), caspase-9 activation, and cytochrome C release to the cytoplasm. These 2 pathways activate the proteolytic cascade and induce cell death. Therefore, MOMP is majorly involved in the intrinsic pathway and regulated by BCL-2 proteins (27, 28).
The protein-protein interactions between Bcl-2 members can regulate apoptosis (29). Mcl-1 from the antiapoptotic Bcl-2 family has shown overexpression in different cancers, including leukemia (27). On the outer mitochondrial membrane, Mcl-1 neutralized the proapoptotic members of the Bcl-2 family, including Bax, Bim, and Bak; in this way, it could block the mitochondrial pathway apoptosis (30). Researchers have demonstrated that Mcl-1 downregulation can result in apoptosis and prevent tumor cell proliferation (31-33).
The suppressive effects of specific siRNA on Mcl-1 proteins in the Jurkat cell line (T-ALL) were evaluated. In this study, quantification of mRNA by qRT-PCR confirmed that Mcl-1 siRNA transfection could significantly reduce Mcl-1 mRNA expression in Jurkat cells. Karami et al. showed that siRNA knockdown of Mcl-1 notably stimulated apoptosis and prevented proliferation in AML (23). In addition, another study on siRNA MCL-1 found it to be a proper target for CLL and ALL; its silencing could significantly increase the therapeutic effect of rituximab (34). The findings of the present study are consistent with previous research on leukemia. The therapeutic method of this study using siRNA could significantly downregulate Mcl-1 mRNA in Jurkat cells. Consequently, inhibition of Mcl-1 expression by siRNA, in combination with other cancer treatments (e.g., immunotherapy with antibodies), could enhance the sensitivity of chemotherapy drugs. However, further studies on combination therapy for leukemia are suggested.
According to the MTT assay, Mcl-1 siRNA had a major contribution to the survival of leukemia cells. Based on the result of this study, the negative control scramble siRNA and reagent caused no changes in gene expression and cellular survival. According to a study by Hao et al. On the human gastric cancer cell, SGC-7901, cell proliferation was examined by MTT assay. The results revealed that Bcl-2 expression knockdown by siRNA reduced gastric cancer cell growth (35). Based on the present study, Mcl-1 siRNA hindered Mcl-1 expression as an antiapoptotic agent in lymphocyte leukemia, and after transfection with specific siRNA, leukemia cell survival decreased.
On the other hand, another study by Meng et al. on pediatric patients with A-BLL demonstrated that transfection of Bcl-2-siRNA into leukemic cells significantly increased the apoptosis rate (36).
Another study in 2013 showed that using Bcl-2 siRNA could significantly increase apoptosis and mortality rate in the gastric cancer cell, BGC823. In addition, they indicated that by treatment with siRNA Bcl-2, a notable increase was observed in the sensitivity of cancer cells to X-ray (37). It is likely that downregulation of Mcl-1 can reduce resistance to X-ray irradiation in the Jurkat cell line. Therefore, further studies are recommended to identify the effects of antiapoptotic proteins on resistance to chemotherapy or radiotherapy.
Moreover, a recent study in 2013 on breast cancer indicated that miR-26a, which is a potential tumor suppressor, inhibited tumor migration, proliferation, and triggered apoptosis via Mcl-1 targeting (38). Similarly, based on the present study, Mcl-1 expression by siRNA could be suppressed and reduce cell survival in leukemia cells. If this method affects tumor migration, further research on tumor migration is needed.
5.1. Conclusions
Collectively, the present study showed that downregulation of Mcl-1 by specific siRNAs can give rise to death in T-ALL cells. Likewise, this study demonstrated that specific siRNAs can be suggested as an effective complementary therapeutic agent for the treatment and management of T-ALL.