1. Background
Breast cancer is the most commonly diagnosed malignancy in women worldwide and is considered to be responsible for 30% of the total new cancer cases and 14% of overall cancer mortality (1). In spite of huge efforts, the definite molecular mechanisms of breast cancer tumorigenesis have largely remained unknown. Therefore, discovering the mechanisms involved in breast cancer and unraveling the factors that contribute to the formation of such mechanisms, will help scientists to define new diagnostic and prognostic biomarkers for this cancer (2, 3). On the other side, it has been shown that three molecular pathways, including cell death, autophagy, and angiogenesis are dysregulated in breast cancer. Biomarkers used for early detection of cancer have the potency to reduce the burden of cancer in the population (4). MicroRNAs (miRNAs) are one of the most important classes of biological molecules and their role in the regulation of various molecular mechanisms has been proven by many researches (5). MiRNA expression profiles are dysregulated in different types of cancers. Some studies have suggested that miRNA expression levels correlate with prognosis of breast cancer (6-8); miR-638 is one of these miRNAs, which is located on chromosome 19p13.2 and downstream of ILF3 and QTRT1 genes (9, 10).
Altered expression of miR-638 has been identified in several different types of human tumors. For example, its expression was dramatically reduced in the human gastric tumor (11), basal cell carcinoma (12), breast cancer (13), non-small cell lung cancer (14), colorectal tumor (15), and chronic lymphocytic leukemia (16); therefore it might have a potential suppressing role in these cancers. Moreover, researchers found that miR-638 has a role in the regulation of the metastasis, autophagy and apoptosis processes in melanoma (17). A recent study demonstrated that the expression level of miR-638 was decreased in tissue samples of liver cancer compared with normal liver tissues, leading to angiogenesis promotion (18).
2. Objectives
The aim of the current study was (a) to use bioinformatics resources to prioritize susceptible miRNAs implicated in breast cancer, (b) to evaluate the expression level of prone miRNAs in clinical samples, (c) to use bioinformatics analysis of candidate miRNAs by referring to different databases in order to detect potential signaling pathways that they are involved in, and (d) to investigate the expression analysis of miR-638 in patients with breast cancer.
3. Methods
3.1. RNA Prediction
At first, the Target Scan (TargetScanHuman7.1) and Diana Tools (Diana Tools MicroT-CDS) were used to predict miR-638 target genes which have the potential binding sites on their three prime untranslated region, according to the Software for Statistical Folding of Nucleic Acids and Studies of Regulatory RNAs (SFold database) (http://sfold.wadsworth.org). Then, the Human Protein Atlas (https://www.proteinatlas.org/) and the Human microRNA Disease Database v3.0 (HMDD v3.0) (http://www.cuilab.cn/hmdd) web-based programs were used to determine mRNA and miRNA expression levels.
3.2. Samples Collection
This study was conducted on a total of 47 breast cancer samples and matched-pair adjacent non-cancerous tissues from newly diagnosed patients with breast cancer at the Imam Khomeini Hospital of Tehran University of Medical Sciences between 2015 and 2018. All the clinical samples were collected from Iran National Tumor Bank (The Cancer Institute, Imam Khomeini Hospital, Tehran, Iran). All the contributors were asked to read and sign an informed consent prior to tumor sample collection and following molecular analysis for the present study.
Specimens were removed during a surgical procedure, and a pathologist distinguished tumor tissue from adjacent healthy tissue, as a normal sample. Then the specimens were immediately frozen in liquid nitrogen, and stored at -80ºC until RNA extraction. All tests were carried out in conformity with relevant guidelines.
3.3. RNA Isolation
Total RNA was extracted from the samples using TRIZOL-Reagent RNA isolation agent (Invitrogen Carlsbad, CA). RNA extraction from tissue samples was performed according to the manufacturer’s protocol. The quantity of RNA was measured using Nano Drop spectrophotometer ND-2000 (Thermo Scientific), and RNA quality was assessed by running the RNA on 1.5% agarose gel electrophoresis.
3.4. cDNA Synthesis and qPCR
Complementary DNA (cDNA) synthesis was performed on 1 µg of extracted total RNA, using a TruScript First Strand cDNA Synthesis Kit (Norgen Canada) for 5srRNA (5S ribosomal RNA; as endogenous control of miRNA) according to manufacturer’s protocol. Quantitative polymerase chain reaction (qPCR) was done in triplicate, for each sample, on the Rotor-Gene Q instrument (Qiagen) using 2X Real-Time PCR Master Mix (Norgen, Canada). 2X SYBR Green master mix (5 µL), 1 µL of cDNA samples, 0.5 µL of each forward and reverse primers (2 pmol) and 3 µL of nuclease-free water (Norgen, Canada) were mingled in Strip Tubes and Caps, 0.1 mL (Qiagen) to perform PCR in a 10 µL reaction volume. The PCR amplifications were performed according to the following program for the thermal cycler instrument: an initial denaturing step for 2 min at 95ºC, followed by 40 cycles of denaturation at 95ºC and combined annealing/extension at 60ºC for 20 sec and 30 sec, respectively. 5srRNA was used as the reference for expression alterations comparison and the fold change in the expression of each target mRNA, relative to 5srRNA, was evaluated based on the 2−ΔΔct comparative expression Livak method (19).
The quantitative assay of mature miRNAs was carried out using the miR-Q method as described by Sharbati-Tehrani et al. (20) with sequence-specific oligonucleotide primers. Table 1 describes the utilized oligonucleotides. Briefly, 3 μL of miRNA cDNA products was added to master mix containing 1 μL MP primers (Fw + Rev), 1 μL short-miR-x and 5 μL of SYBR, in total volume of 10 μL. The qPCRs were executed in triplicate according to the standard program on Rotor-Gene Q instrument (QIAGEN, Germany). Thermal cycler was programmed to run according to the following conditions: 95ºC for 2 min followed by 40 cycles of denaturing at 95ºC for 15s and combined annealing-extension for 30 s at 60ºC. Moreover, a non-template control (NTC) reaction was employed for each primer set to guarantee the reaction specificity.
| Name | Sequence (5’ to 3’) |
|---|---|
| 5S rRNA- Fw | GCCCGATCTCGTCTGATCT |
| 5S rRNA-Rev | AGCCTACAGCACCCGGTATT |
| RT6-miR-638 | TGTCAGGCA ACCGTATTCACCGTGAGTAGGCCG |
| Short-miR-638 | CGTCAGATGTCCGAGTAGAGGGGGAACGGCAGGGATCGCGGGCGGGTGG |
| MP-Fw | TGTCAGGCAACCGTATTCACC |
| MP-Rev | CGTCAGATGTCCGAGTAGAGG |
Designed Oligonucleotides for the Target Sequences
3.5. Bioinformatics Analysis
The study was carried out through bioinformatics methods and by using miRWalk, MicroRNA-Target Interactions (miRTarBase) database, and the database for annotation, visualization and integrated discovery (DAVID). MiRWalk2.0 database, that predicts miRNA-mRNA interactions, provided a list of more than a thousand genes. By using miRTarBase, prone genes were evaluated and compared with the previous list.
Then, by using the UniGene database, the expression of target mRNAs was analyzed in mammary gland cells to determine which mRNA is expressed in the mammary gland. Ultimately, to carry out signaling pathways enrichment analysis, miR-638 targetome expressed in mammary glands was included in the DAVID database 6.8. This database yields results obtained from pathway analyses performed by the Kyoto Encyclopedia of Genes and Genomes (KEGG) database to detect the molecular networks and signaling pathways including the most miR-638 targetome.
3.6. Statistical Analysis
The data obtained from real-time PCR were analyzed by using the ΔΔCT method in SPSS software version 25.0 (Chicago, IL, USA). Normal distribution of data was examined by the Shapiro-Wilk test. All tests were carried out in triplicate, and data were presented as mean [± standard error of measurement (SEM)]. Wilcoxon and Mann-Whitney U tests were used for analysis with SPSS (version 25.0) and GraphPad Prism (version 6.0) software. In all the tests, P value < 0.05 indicated a significant difference.
4. Results
4.1. Characteristics of Collected Samples
The study consisted of 47 breast tumor specimens and their paired non-cancerous samples. Information regarding clinicopathological factors and demographic characteristics such as age, tumor size, grade, stage, site of primary tumor, necrosis presence, vascular invasion, perineural invasion, family history, lymphatic invasion, and ductal carcinoma in situ (DCIS) have been described in Table 2. From the point of the tumor size, stage and grade, the majority of the tumors were less than 5 cm, stage 2 and grade 2, respectively. Most of the patients didn’t have a positive family history; however, their tumors had a vascular and lymphatic invasion.
| Variable | Value |
|---|---|
| Tumor size (%) | |
| < 3 cm | 17 (36.2) |
| ≥ 3 to < 5 cm | 17 (36.2) |
| ≥ 5cm | 13 (27.7) |
| Grade (%) | |
| I | 4 (8.5) |
| II | 22 (46.8) |
| III | 21 (44.7) |
| Stage (%) | |
| II | 32 (68.1) |
| III | 15 (31.9) |
| Site of primary tumor | |
| Right Breast | 25 (53.2) |
| Left Breast | 18 (38.3) |
| Bilateral | 4 (8.5) |
| Necrosis presence | |
| Yes | 21 (44.7) |
| No | 26 (55.3) |
| Vascular invasion | |
| Yes | 30 (63.8) |
| No | 17 (36.2) |
| Perineural invasion | |
| Yes | 15 (31.9) |
| No | 32 (68.1) |
| Family history | |
| Yes | 15 (31.9) |
| No | 32 (68.1) |
| Lymphatic invasion | |
| Yes | 27 (57.1) |
| No | 20 (42.6) |
| DCIS | |
| Present | 36 (76.6) |
| Absent | 11 (23.4) |
General Demographic Data of Study Participants
4.2. Expression of miR-638 in Breast Cancer Samples
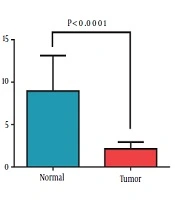
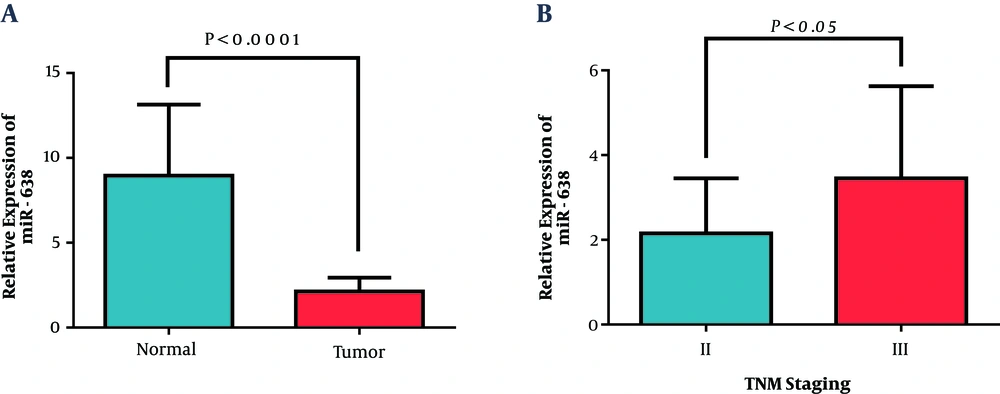
Data showed that the expression of miR-638 in tumoral tissues decreased significantly, in comparison with their paired normal adjacent tissues (2.13 ± 0.82). Figure 1A shows the relative miR-638 expression in patients and the control group. Correlation analysis was conducted to find possible relations between the expression of miR-638 and the clinicopathological characteristics of the patients, including age, site of primary tumor, tumor size, grade, stage, and HER2 status (Table 3). A significant low expression of miR-638 was observed in the tumoral samples, in comparison with adjacent non-tumoral samples. Interestingly, with the progression of disease from stage 2 to stage 3, the expression level of miR-638 increased, which implicated cancer growth. As shown below, the mean expression of miR-638 in the stage II group was 0.67 ± 0.12, while in low tumor stage cluster was 1.14 ± 0.20 (P = 0.041; Figure 1B). Nevertheless, no significant association was observed in respect to age, site of primary tumor, tumor size, grade, and HER2 (Table 3).
Expression of miR-638 was evaluated by RT-qPCR in breast cancer tissues. A) According to the statistical analysis, miR-638 expression decreased in breast cancer tissue samples, compared with adjacent noncancerous tissue samples (Wilcoxon test). B) Differential expression of miR-638 in stage II and III of breast cancer patients. Values represent means and error bars represent the SEM (P < 0.0001 and P < 0.05 respectively).
| Feature | miR-638 | |
|---|---|---|
| Mean ± SEM | P | |
| Age | ||
| < 54 | 0.72 ± 0.15 | 0.196 |
| ≥ 54 | 0.93 ± 0.16 | |
| Site of primary tumor | ||
| Left | 0.98 ± 0.18 | 0.277 |
| Right | 0.69 ± 0.14 | |
| Bilateral | 0.98 ± 0.48 | |
| Tumor size | ||
| < 3 cm | 0.83 ± 0.19 | 0.928 |
| ≥ 3 to < 5 cm | 0.78 ± 0.17 | |
| ≥ 5cm | 0.19 ± 0.22 | |
| Grade | ||
| I | 1.01 ± 0.48 | 0.655 |
| II | 0.73 ± 0.16 | |
| III | 0.89 ± 0.15 | |
| Stage | ||
| II | 0.67 ± 0.12 | 0.041 |
| III | 1.14 ± 0.20 | |
| HER2 | ||
| Positive | 1.18 ± 0.21 | 0.167 |
| Negative | 0.74 ± 0.15 | |
Association of miR-638 Expression Levels and Clinicopathological Characteristics of Cancerous Breast Samples
4.3. Signaling Pathway Enrichment Analysis of miR-638 Targetome
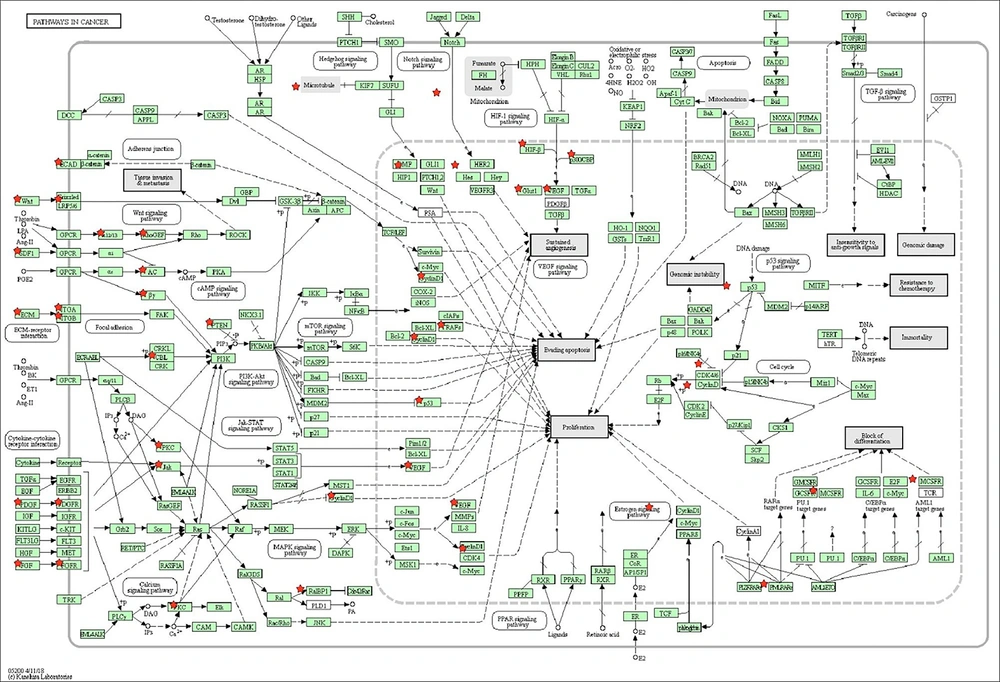
Molecular signaling pathway enrichment analysis was conducted to study the potential roles of miR-638 in the tumorigenesis and progression of breast cancer. All of the 1152 predicted mRNAs and 53 validated mRNAs which were obtained from miRWalk and miRTarBase respectively were targeted by miR-638. All the validated targets drawn from the miRTarBase database were confirmed by persuasive tests, such as western blot, quantitative real-time PCR, and reporter assay. Thirty-eight out of the 53 validated mRNAs (71%) were also observed in the list of predicted mRNAs, confirmed by at least five prediction databases, which demonstrated that the threshold of five databases is reliable to select predicted miR-638 targets. The targets’ expression profiles in the UniGene database revealed that only 38 out of the validated mRNA targets and 1018 out of the predicted ones were expressed in mammary gland cells. These targets were selected as miR-638 targetome for additional molecular enrichment analysis. Next, the selected miR-638 targetome was entered into the DAVID functional annotation to investigate whether the association of the entered genes with signaling pathways and biological terms was statistically significant. Findings from the miRWalk2.0 database and signaling pathways, which have been obtained from DAVID, indicated that the change in the expression level of miR-638 could deviate the cell from its original pathway by affecting cells’ target genes during apoptosis, autophagy, and angiogenesis (Figure 2). Table 4 displays the pathways obtained from the DAVID database and their P value.
| KEGG Pathway | P Value |
|---|---|
| MicroRNA in cancer | 4.3E-6 |
| PI3K-Akt signaling pathway | 5.7E-4 |
| Notch signaling pathway | 7.7E-4 |
| Pathway in cancer | 2.1E-3 |
| AMPk signaling pathway | 2.2E-3 |
The Signaling Paths Obtained from the DAVID Database and Their P-Value Derived from DAVID
5. Discussion
Recent studies have indicated that miRNAs play a crucial regulatory role in many cancers (21). Therefore, it can be deduced that miRNAs may have a promising and prominent prognostic and diagnostic value in different cancers, such as breast cancer (22). The data obtained from the present study showed that the expression level of miR-638 was down-regulated in breast cancer tissues compared to matched-tumor-adjacent tissues. Furthermore, reduced miR-638 expression was correlated with clinicopathological characteristics of breast cancer patients, including advanced stage. The expression level of miR-638 was higher in stage 3, compared with stage 2 (Figure 1B). Since the down-regulation of miR-638 occurred in lower stages, but not in higher stages, it may indicate that the need for more angiogenesis is the cause of the up-regulation of miR-638. From obtained results and bioinformatics data, it can be inferred that the low expression of miR-638 is associated with a decrease in apoptosis. Thus, it can be understood that the up-regulation of miR-638 in patients with breast cancer, is associated with poor prognosis; for this reason, this miRNA can be used as a potential prognostic biomarker.
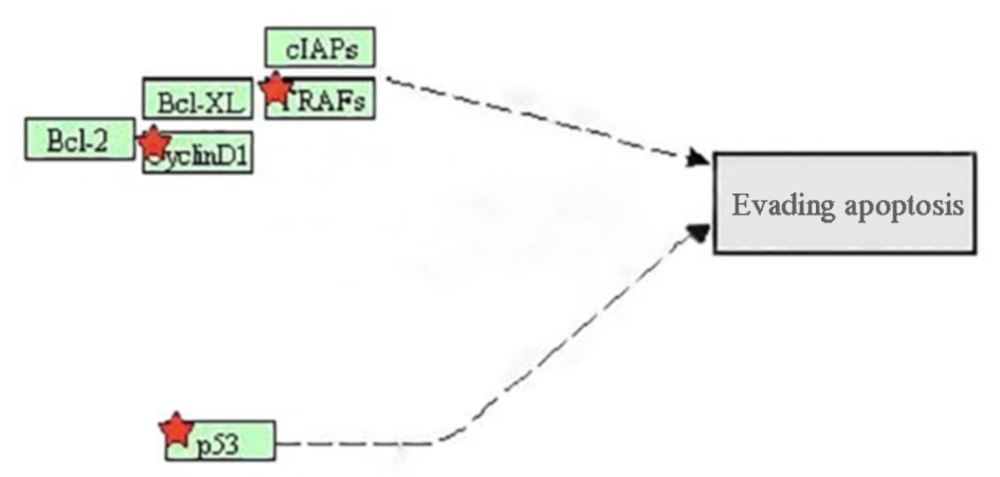
Bioinformatics analysis revealed that dysregulation of miR-638 had a role in angiogenesis and the inhibition of autophagy. miR-638 has many target genes including ATG-5, ATG-2B, VEGF, TP53, and TRAF (23, 24). Apoptosis is a normal cellular process which leads to controlling the growth and development rate. Excessive activity of apoptosis causes diseases such as Alzheimer’s and Parkinson and if apoptosis rate is too little, it can lead to cancer (25). The genes involved in apoptosis have the potential to be regulated by microRNAs. In the apoptosis pathway, which was induced by miR-638, a change in the P53 protein was observed. TP53 is a tumor suppressor gene and mutations in this gene have been indicated in most of the human cancers. This gene plays an important role in suppressing the tumor, stopping the cell cycle, inducing apoptosis, and inhibiting angiogenesis (26).
The proteins that play a role in apoptosis pathway are divided into 2 groups: (i) anti-apoptotic proteins, which include Bcl-2 and TRAFs, and (ii) pro-apoptotic members that include BAX, and BAK. These proteins, by binding to the outer membrane of mitochondria and inhibition of pro-apoptotic family members, maintain the mitochondrial membrane and inhibit apoptosis (27) (Figure 3).
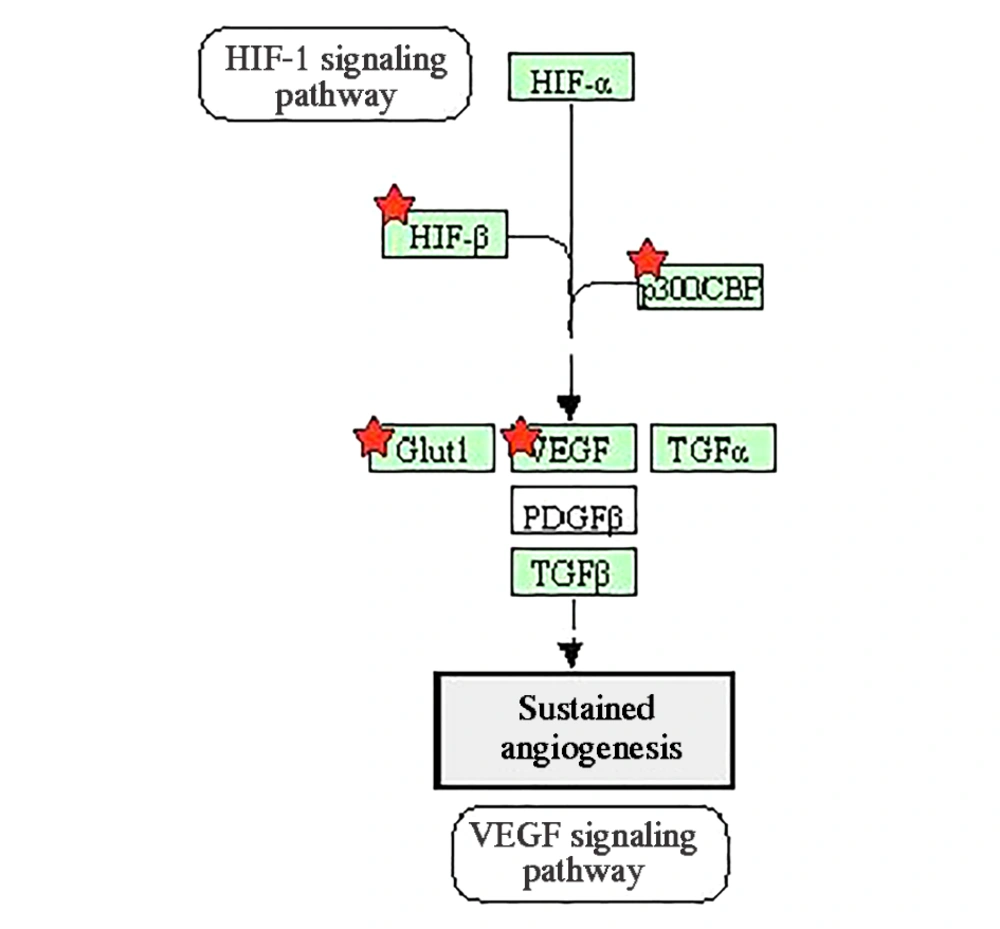
Since tumor cells have fast and uncontrolled growth in comparison to normal cells, some of them that are far from blood vessels encounter with hypoxia or lack of oxygen. Therefore, angiogenesis begins inside and around the tumor to provide the oxygen needed for the cancer cells (28). VEGF is one of the most significant proteins in the angiogenesis process, and miR-638 alters its expression (18). Figure 4 shows the genes induced by miR-638 in the angiogenesis pathway.
Bioinformatics analyses indicated that miR-638 exerts its effect through affecting ATG-5 and ATG-2B proteins, which play a role in cellular autophagy. MiR-638 leads to autophagy inhibition by targeting the genes in the elongation phase. Further studies are needed to illustrate the effects of reducing the expression of miR-638 in autophagy. The findings of this study indicate how miR-638 affects intracellular pathways, including autophagy and angiogenesis, which lead to the progression of breast cancer.
Our results and bioinformatics analysis showed that downregulation of miR-638 may be associated with a good prognosis in breast cancer, because with decreases in miR-638 expression level, we observed the overexpression of ATG-5 and TP53 genes, which are involved in autophagy and apoptosis, respectively.
In line with our study, Zhang et al. observed that miR-638 expression was reduced in colorectal cancer tissues, in comparison to their adjacent normal tissues (15). On the other hand, the mean expression level of miR-638 was significantly up-regulated in prostate and kidney cancers (29). Taken together, it can be suggested that miR-638 may have a potential prognostic role in breast cancer patients.
5.1. Conclusions
The results of the present study indicate a significant expression pattern difference of miR-638 in breast tumor specimens and their paired non-cancerous samples. Our findings showed that miR-638 was down-regulated in cancerous samples and was associated with higher stages. We conducted bioinformatics research to reach the target genes of miR-638, which can have the greatest impact on the pathways leading to cancer progression, and the possibility of using this miRNA in cancer prognosis. These results demonstrate that miR-638 might serve as a potential prognostic biomarker; however, more investigation is needed.