1. Background
Breast cancer is the most frequently diagnosed cancer among women accounting for about one-third of all new cancers in women (1). The most common type of breast cancer is invasive ductal carcinoma, which accounts for 50% to 80% of all cases (2). Most breast cancers have been shown to be hormone receptor positive with 75% of them being estrogen receptor (ER) positive and 55% being progesterone receptor (PR) positive (3). Although several treatment modalities have been designed for patients with breast cancer based on the expression of these markers, a significant number of patients do not benefit from these treatment strategies. Based on the heterogeneous nature of breast cancer, which incorporates numerous subgroups with variable molecular profiles, prognoses, and responses to therapies (4), several groups have aimed at identifying molecular targets with potentials to be used in therapeutic strategies. Although mRNA coding genes have been presented as special biomarkers (5-8) and therapeutic targets (9-12) for several years, non-coding RNAs have just recently attained attentions of researchers. A group of these transcripts, called as long non-coding RNAs (lncRNAs), have the length of more than 200 nucleotides, which differentiates them from classic non-coding RNAs, such as ribosomal (r)RNAs, ribozymes, transfer (t)RNAs, and microRNAs (13). In addition, they have been shown to participate in many central biologic processes comprising regulation of gene expression, telomere length, chromatin reorganization, histone alterations, genomic imprinting, and cell differentiation all of them being involved in cancer development (14, 15). We have previously evaluated the expression of a number of lncRNAs in breast cancer in relation to patients’ clinical characteristics to determine the role of these transcripts in breast tumorigenesis (16, 17). In the present study, we aimed at identifying lncRNA-gastric carcinoma high expressed transcript 1 (lncRNA-GHET1, AK123072) expression in breast cancer tissues compared with their adjacent noncancerous tissues (ANCT) in a cohort of Iranian patients. As its name implies, GHET1 has been firstly characterized as an lncRNA over-expressed and participated in the pathogenesis of gastric cancer (18). Subsequently, its contribution in the pathogenesis of bladder cancer has been revealed (19). However, to the best of our knowledge, there is no data regarding its expression in breast cancer tissues and its contribution in breast tumorigenesis.
2. Methods
2.1. Tissue Samples
The Ethical Committee of Shahid Beheshti University of Medical Sciences (Tehran, Iran) approved this study. Forty-seven samples of invasive ductal carcinoma of breast, in addition to their ANCTs, were obtained from patients in Sina and Farmanieh hospitals under the conventions of the ethics committee. ANCT was defined as the normal breast tissue and the diagnosis was confirmed by the pathologists through H.E. staining. Informed consent was obtained from all participants. The clinical and pathological data of patients were gathered through questionnaires and evaluation of medical records. The samples were instantly snap-frozen in liquid nitrogen before being stored in -70°C.
2.2. RNA Extraction and Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction (RT-PCR)
The AccuZolTM total RNA extraction solution (Bioneer, Korea) was used for RNA extraction according to the manufacturer’s instructions. A 1 hour treatment with DNase I has also been included. The purity, concentration, and integrity of RNA were analyzed by Thermo Scientific NanoDropTM 1000 Spectrophotometer and gel electrophoresis, respectively. Then, cDNA was synthetized from all samples, using 1µg of RNA in PrimeScript RT reagent kit (Takara Bio, Ohtsu, Japan). Synthesized cDNA was, then, checked spectrophotometrically to appraise its concentration. Quantitative RT-PCR reaction was performed on a rotor gene 6000 corbette detection system, using SYBR Premix Ex Taq (Takara Bio, Ohtsu, Japan). Thermal cycling conditions were an initial activation step for 5 minutes at 95º C followed by 40 cycles at 95°C for 15 seconds, specific annealing temperature for 15 seconds, and 72°C for 20 seconds. The specific annealing temperatures were 63°C for GHET1 and B2M and 56.5°C for MYC. No template control (NTC) containing H2O was included in each run. B2M gene was used as normalizer. Forward and reverse primers sequences are as follows, respectively: GHET1: 5’- AGTCAGCTCCCTACAGAGGTG-3’ and 5’-TCCTTAGGTGGTGGTTTCTGTTC-3’; B2M: 5’-AGATGAGTATGCCTGCCGTG-3’ and 5’-GCGGCATCTTCAAACCTCCA-3’; MYC: 5’-CACATCAGCACAACTACG-3’ and 5’-GTTCGCCTCTTGACATTC-3’. Melting curve analysis was carried out to validate specificity of PCR products. In addition, PCR products were electrophoresed on 2% agarose gel to verify product sizes and specificity.
2.3. Estrogen Receptor (ER), Progesterone Receptor (PR), Her2/neu, and Ki-67 Status
Patients’ medical records were used for evaluation of these markers. In brief, all of these markers were assessed by immunohistochemical (IHC) staining. For ER and PR, staining of > 20% of tumor cell nuclei was considered positive. For Her2/neu, a test result of 0 to 2+ was regarded as negative and 3+ as positive. Ki-67 values were reported as both the percentage of positively stained malignant cells among the entire number of malignant cells and alternatively as positive vs. negative.
2.4. Statistical Analysis
Fold changes in gene expression were calculated by LinRegPCR (2) and relative expression software Tool-RG©-version 3 (QIAGEN, Korea). The amplification efficiencies and cycle thresholds were used for such comparative quantification analysis. The quantities of mRNAs in the samples were normalized to the B2M mRNA and compared between paired tumor and ANCTs. The significant alterations in gene expression were determined by the pairwise fixed reallocation randomization test with 2000 iterations in the REST 2009 software. The level of statistical significance was set at P < 0.05.
Statistical analyses were executed by SPSSv.20 (SPSS Inc., Chicago, IL). The Wilcoxon signed-rank and McNemar tests were used when comparing paired tumor and ANCTs. Chi-square and independent t tests were used for testing the relationship between categorical variables. Significance was defined as P < 0.05.
3. Results
3.1. General Statistical Information
Breast tissues from a total of 47 female patients with breast cancer with the mean age of 51.35 ± 14.721 (23 - 84) were drawn from our previous study (16) and recruited in the current project. Clinicopathological data such as patients’ age, tumor size, tumor stage and grade, ER, PR, Her2/neu, and Ki67 status have been investigated based on the evidences acquired from questionnaires, interviews, and clinical and laboratory tests as mentioned in the previous study (16).
| Characteristics | Down Regulation | Up Regulation | No. | P Value |
|---|---|---|---|---|
| Age | 0.2 | |||
| < 50 | 17 | 8 | 25 | |
| ≥ 50 | 17 | 4 | 21 | |
| Stage | 0.147 | |||
| 0 | 1 | 0 | 1 | |
| I | 3 | 1 | 4 | |
| II | 19 | 7 | 26 | |
| III | 11 | 2 | 13 | |
| IV | 0 | 2 | 2 | |
| Histological grade | 0.256 | |||
| I | 4 | 2 | 6 | |
| II | 20 | 4 | 24 | |
| III | 9 | 6 | 15 | |
| Family history | 0.259 | |||
| No | 23 | 10 | 33 | |
| Yes | 11 | 2 | 13 | |
| Tumor size, cm | 0.194 | |||
| < 2 | 7 | 1 | 8 | |
| ≥ 2, < 5 | 25 | 9 | 34 | |
| ≥ 5 | 1 | 2 | 3 | |
| Node status | 0.618 | |||
| Negative | 20 | 7 | 27 | |
| Positive | 14 | 5 | 19 | |
| ER status | 0.641 | |||
| Negative | 9 | 24 | 33 | |
| Positive | 3 | 8 | 11 | |
| PR status | 0.435 | |||
| Negative | 12 | 3 | 15 | |
| Positive | 21 | 8 | 29 | |
| Her2/neu status | 0.516 | |||
| Negative | 25 | 9 | 34 | |
| Positive | 8 | 2 | 10 | |
| Ki67 status | 0.578 | |||
| Negative | 2 | 1 | 3 | |
| Positive | 29 | 9 | 38 | |
| Hormone replacement therapy | 0.245 | |||
| Negative | 26 | 11 | 37 | |
| Positive | 8 | 1 | 9 | |
| Body mass index | 0.012 | |||
| 18.5 - 24.9 | 0 | 3 | 3 | |
| 25 - 29.9 | 20 | 5 | 25 | |
| > 30 | 13 | 4 | 17 | |
| Smoking history | 0.162 | |||
| Negative | 33 | 10 | 43 | |
| Positive | 1 | 2 | 3 |
LncRNA-GHET1 Expression and its Associations with Patients’ Clinical and Demographic Data
3.2. Expression of GHET1 and MYC in Patients’ Samples
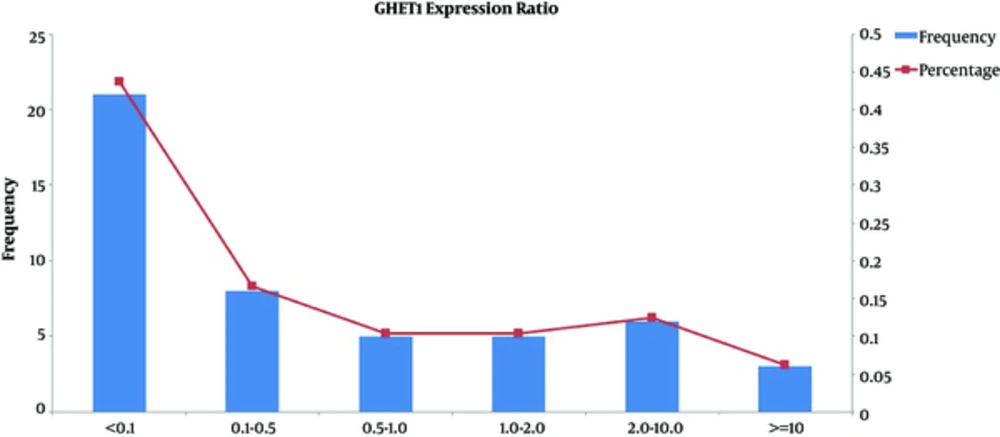
Significant elevated GHET1 expression was identified in 13/47 (26%) of cancer tissue samples compared with their corresponding ANCTs. However, the comparison of GHET1 transcript levels between total tumor and ANCT tissues demonstrated a significant down-regulation in tumor tissues compared with ANCTs (P = 0.02). Figure 1 shows the frequency and percentage of samples in certain subgroups based on relative expression of GHET1 in tumor tissues compared with ANCTs. No significant association has been found between the level of GHET1 transcripts and those of MYC (P = 0.324).
3.3. Correlations Between GHET1 Expression and Clinical Characteristics
To further determine the role of GHET1 in breast cancer, we subsequently analyzed the associations between its transcript levels and numerous clinicopathological characteristics. However, no significant associations have been found between GHET1 expression and these features except for an association between GHET1 transcript level and body mass index. The relationship between GHET1 expression and clinicopathological variables is demonstrated in Table 1. Then, the patients were divided into 3 groups based on the relative GHET1 expression (REX) in tumor tissues compared with the corresponding ANCT: over-expressed (REX > 1, n = 13), moderately down-regulated (0.185 < REX < 1, n = 8), and highly down-regulated (REX ≤ 0.185, n = 26). The average level of REX in patients with GHET1 down-regulation was 0.185, so this value was chosen for further categorization of patients. Statistical analyses performed for these 3 groups showed no significant association between GHET1 transcript levels and clinicopathological data. As the expression analyses have been performed on the same samples as our previous study, we further analyzed the associations between GHET1 expression and 2 other lncRNAs, whose expressions have been evaluated in the same samples previously by our group (16). A significant association has been found between GHET1 and CCAT2 expression (P = 0.038), while no association was detected between the levels of GHET1 and aHIF transcripts (P = 0.415).
4. Discussion
In the present study, for the first time we demonstrated GHET1 over-expression in 26% of samples with breast invasive ductal carcinoma relative to their paired ANCTs. Previous studies have shown many lncRNAs to be dysregulated during breast tumorigenesis. According to their expression pattern in tumor tissues and their putative function, they have been categorized into tumor suppressor and oncogene lncRNAs (14). Formerly, GHET1 has been shown to enhance the proliferation of gastric carcinoma cells in vitro and in vivo. Its function is probably mediated by increasing c-Myc mRNA stability and expression (18). In addition, GHET1 over-expression has been demonstrated in bladder cancer tissues compared to adjacent normal tissues and its transcript levels were correlated with tumor size, advanced stages, lymph node involvement, and poor prognosis. Its role in bladder cancer pathogenesis has been confirmed by loss-of- function analysis, which showed that it is involved in the proliferation and invasion of bladder cancer cells via regulation cell cycle checkpoints as well as the epithelial-mesenchymal-transition (19). Consequently, GHET1 can be regarded as an oncogene in these 2 types of cancer. In addition, GHET1 have been shown to directly interact with insulin-like growth factor 2 mRNA binding protein 1 (IGF2BP1) and have increased the physical interface between MYC mRNA and IGF2BP1 (18). Considering the role of GHET1 in regulation of MYC mRNA stability and expression via its interaction with IGF2BP1 (18), frequent MYC dysregulation in breast cancer (20) as well as expression of IGF2BP1 in breast cancer and its role in clonogenic growth of these cells (21), GHET1 was expected to participate in breast cancer pathogenesis as well. However, the results of the present study showed no consistent expression trend in samples with breast cancer examined. Furthermore, its transcript levels were neither correlated with prognostic markers nor with the MYC transcript levels. Future studies with larger sample sizes are needed to evaluate its significance in breast tumorigenesis. As the majority of non-protein-coding transcripts demonstrate cell-specific expression patterns (22), their putative role on the pathogenesis of distinct cancers might be different. However, there are certain lncRNAs, which have a similar function in the development of diverse cancers (14, 15). On the other hand, although we demonstrated the dysregulation of GHET1 in breast cancer tissues, further functional analyses are needed to evaluate its precise role during breast tumorigenesis. Based on the genetic and epigenetic instabilities detected in tumoral tissues, it is possible that dysregulation of expression of many genes in these tissues are the simple by-product of stochastic situation caused by such instabilities. Consequently, in vitro loss-of-function and gain-of-function studies would facilitate the determination of GHET1 function during breast tumorigenesis.
4.1. Conclusions
Although previous studies have demonstrated GHET1 over-expression in gastric and bladder cancers and its contribution in the pathogenesis of both cancers, in the present study, we could detect its over-expression in only 26% of breast tumor samples.