1. Background
Atrial fibrillation (AF), the most prevalent supraventricular arrhythmia, affects up to 1% of the general population, with the prevalence increasing to 10% in the elderly (1). It is estimated that more than 33 million people worldwide suffer from AF, with the incidence being three times higher in men compared to women (2). The incidence and outcomes of AF are influenced by many genetic and epigenetic factors and vary significantly by geography and ethnicity (3). Electrocardiography (ECG) is the most common method used for AF diagnosis. However, due to its short recording duration, detecting AF in asymptomatic patients is limited. Therefore, useful biomarkers are needed for the early identification and treatment of asymptomatic AF cases (4).
Several general risk factors, such as diabetes, heart failure, hyperthyroidism, arterial hypertension, obesity, gender, and ischemic and structural heart disease, are known to be associated with AF. In more than 20% of AF subjects, there is no evidence of any underlying risk factors, which are known as lone AF (5). It has been proven that individuals with at least one parent with a history of AF are more susceptible to developing AF, implying the role of genetic factors in the pathogenesis of AF (6). However, a large part of the heritability of AF remains unexplained (7).
Micro-RNAs (miRNAs) are single-stranded, non-protein-coding RNAs containing around 22 nucleotides that regulate the expression of target genes post-transcriptionally by binding to the 3'UTR of mRNA molecules (8). Several processes, including cell growth, differentiation, proliferation, and metabolism, are regulated by miRNAs (9). miRNAs have been commonly detected in circulating blood and cardiac muscle of humans and animal models (10). Additionally, miRNAs have been shown to be associated with the pathophysiology of several cardiovascular diseases, including AF, by regulating atrial remodeling mechanisms (11, 12). Changes in miRNA expression levels have been reported to modulate the susceptibility to AF development in humans. Hence, miRNAs have the potential to be considered targets for the diagnosis and treatment of AF (13).
Changes in miRNA expression have been reported as potential prognostic and diagnostic biomarkers (14). The high stability, sensitivity, specificity, and accessibility of circulating miRNAs in serum and plasma make them interesting targets for the early diagnosis of several cardiac diseases, including AF (15-18).
Considerable efforts have been made to identify plasma markers for AF diagnosis. Several studies have identified circulating microRNA levels and aberrant expression of microRNAs in AF tissue, paving the way for further analysis of circulating microRNAs as biomarkers for AF prediction (19-21).
2. Objectives
Therefore, the aim of this investigation was to explore the expression levels of miR-199a and miR-126 in lone AF patients and to evaluate their diagnostic power for the discrimination of AF.
3. Methods
In the present case-control investigation, 70 participants were included, of which 35 cases had lone AF and 35 were healthy controls. Individuals with lone AF were selected from those referred to Modarres Hospital in Tehran (2022 - 2023) and were subjected to ECG and 24-hour Holter monitoring due to suspected clinical symptoms of AF. Control subjects were selected from those who underwent ECG due to discomfort in the chest, feeling of tachycardia, or heart palpitations, but had a normal sinus rhythm and absence of any arrhythmia.
Patients with inflammatory, infectious, or autoimmune diseases, a history of hematological disease, malignancy, kidney, liver, or thyroid disease, hypertension, diabetes, valvular disease, or a history of previous MI were excluded from the study. This study was performed in line with the principles of the Declaration of Helsinki and according to the accepted regulations of the Shahid Beheshti University of Medical Sciences ethical committee (IR.SBMU.MSP.REC.1402.565). Informed consent was obtained from all participants.
3.1. Genes Selection
The prioritization and selection of the most appropriate genes in the molecular pathways involved in the pathogenesis of lone AF were conducted by collecting and downloading data on the inflammatory pathways of lone AF pathogenesis from KEGG, Reactome, BioCarta, WikiPathways, and previous studies.
3.2. Expression Analysis
Five milliliters of whole blood were taken from all subjects, and the total RNA was isolated from the blood specimens using the RiboEx™ RNA purification kit (GeneAll Biotechnology Co. Ltd, Seoul, Korea). The integrity and concentration of RNA samples were assessed using the Nanodrop instrument (Thermo Fisher Scientific). Complementary DNA (cDNA) synthesis was accomplished using the Applied Biosystems High‐Capacity cDNA Reverse Transcription Kit (Takara, Tokyo, Japan). The expression levels of the miR-199a and miR-126 genes in the RNA samples were assessed using the RealQ Plus 2x Master Mix Green, High ROXTM (AmpliQon, Denmark) on the LightCycler 96 Real-Time PCR System (Roche, Germany). RNU6 was used to normalize the expression results via the 2 -ΔΔCt method. Sequences of specific primers for investigating the expression levels of miR-199a, miR-126, and RNU6 were designed using Allele ID software and are provided in Table 1.
| Gene of Interest | Primer Sequence |
|---|---|
| RNU6 | |
| Forward | 5’-GCTTCGGCAGCACATATACTAAAAT-3’ |
| Reverse | 5’-CGCTTACGAATTTGCGTGTCAT-3’ |
| miR-126 | |
| Forward | 5’-GGGCATTATTACTTTTGG-3’ |
| Reverse | 5’-TGCGTGTCGTGGAGTC-3’ |
| miR-199a | |
| Forward | 5’-ACACTCCAGCTGGGCCCAGTGTTCAGACTAC-3’ |
| Reverse | 5’-CTCAACTGGTGTCGTGGAGTCGGCAA-3’ |
3.3. Statistical Analyses
All statistical analyses were performed using PRISM version 8 and SPSS version 24 statistical software. Continuous variables were described as mean ± standard deviation (SD), and categorical variables were presented as absolute frequency and frequency percentage. The Fisher’s Exact Test was used to compare categorical variables between patients and control subjects. The t-test was applied for continuous variables after verifying the normality of the distribution using the Shapiro-Wilk test. ROC curve analysis of miR-126 and miR-199a expression levels for distinguishing lone AF patients from control subjects was performed using MedCalc software. A p-value of less than 0.05 was considered significant.
4. Results
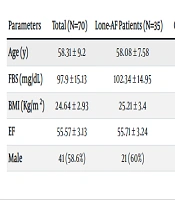
A total of 70 individuals with an average age of 58.31 ± 9.2 years were included in the present study, of whom 58.6% were male. The comparison between lone AF patients and control subjects revealed significant differences only in FBS levels, with lone AF patients having significantly higher FBS levels. The basic and demographic features of the study participants are provided in Table 2.
| Parameters | Total (N=70) | Lone-AF Patients (N=35) | Control Subjects (N=35) | P-Value |
|---|---|---|---|---|
| Age (y) | 58.31 ± 9.2 | 58.08 ± 7.58 | 58.54 ± 10.69 | 0.83 |
| FBS (mg/dL) | 97.9 ± 15.13 | 102.34 ± 14.95 | 93.45 ± 14.16 | 0.01 |
| BMI (Kg/m2) | 24.64 ± 2.93 | 25.21 ± 3.4 | 24.08 ± 2.28 | 0.1 |
| EF | 55.57 ± 3.13 | 55.71 ± 3.24 | 55.42 ± 3.06 | 0.7 |
| Male | 41 (58.6%) | 21 (60%) | 20 (57.1%) | 0.99 |
a Values are expressed as No. (%) or mean ± SD.
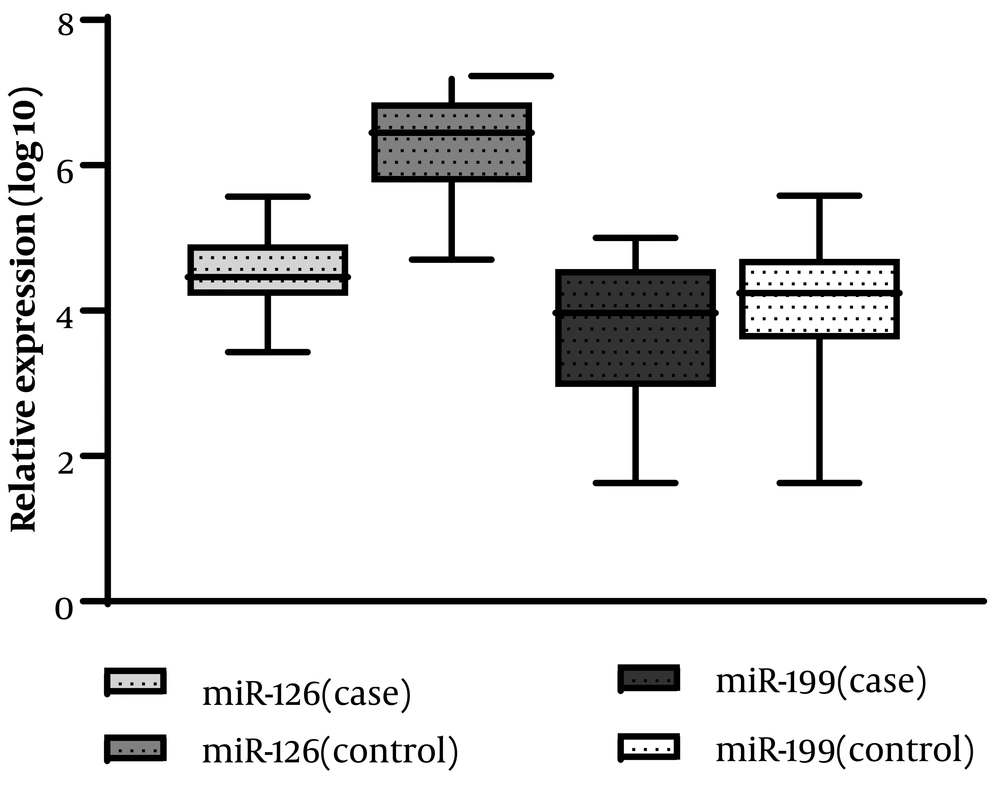
As shown in Figure 1 and Table 3, the relative expression levels of the two investigated miRNAs were compared between lone AF patients and control subjects. The expression analysis revealed that the expression level of miR-126 in patients with lone AF was significantly lower than in healthy controls (P = 0.0001).
a P-value less than or equal to 0.05 is considered significant.
b The expression ratio (fold change) was calculated by the 2-ΔΔct formula.
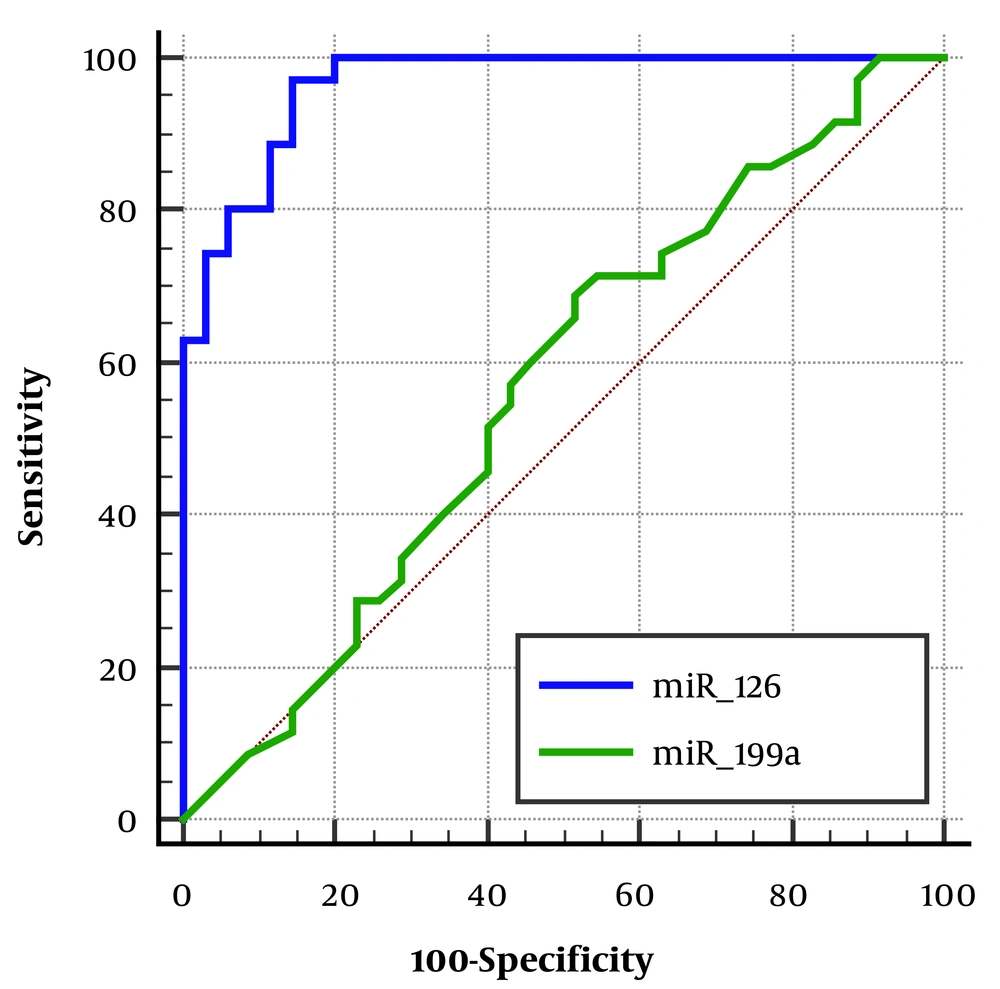
Finally, the diagnostic power of miR-126 and miR-199a expression levels in patients with lone AF was evaluated (Table 4). The expression level of miR-126 showed excellent diagnostic power for the occurrence of lone AF with an AUC of 0.96, while the expression level of miR-199a did not demonstrate significant predictive power (Figure 2).
| Variables | AUC | 95% CI | P-Value | Sensitivity | Specificity | Youden Index (Threshold) |
|---|---|---|---|---|---|---|
| miR-126 | 0.96 | 0.89 to 0.99 | 0.0001 | 97.14 | 85.71 | 0.82 |
| miR-199a | 0.56 | 0.44 to 0.68 | 0.33 | 68.57 | 48.57 | 0.17 |
5. Discussion
Based on the results of this study, the expression level of miR-126 in patients with lone AF was significantly lower than in healthy controls, while the expression level of miR-199a was not significantly different between the patients and control subjects. Additionally, the expression level of miR-126, with an AUC of 0.96, showed excellent predictive power for the occurrence of lone AF, while the expression of miR-199a did not have significant predictive power in differentiating lone AF patients from control subjects.
The molecular mechanisms involved in the early progression of AF are still poorly understood. So far, only a limited number of studies have explored the role of miRNAs as a genetic component in the pathogenesis of AF (20, 22). These investigations typically involved patients with chronic AF, where extensive atrial remodeling processes have already been established. Therefore, changes in the miRNA profile of patients with chronic AF may result from such remodeling processes. In contrast, this study focused on lone AF and revealed a specific miR-protein interaction that may be involved in the early pathogenesis of AF without causing confounding structural changes (23, 24).
These results showed that miR-126 was significantly downregulated in blood samples from patients with lone AF. miR-126, also known as angiomiR-126, acts as an important regulator in maintaining endothelial homeostasis and vascular integrity. Circulating miR-126 has the potential to be used as a biomarker for vascular injury and cardiac damage in cardiovascular diseases (CVDs) including coronary artery disease (CAD), myocardial infarction (AMI), heart failure, and AF (25-28). Decreased circulating levels of miR-126 were associated with endothelial dysfunction (29). The expression level of miR-126 was significantly decreased in persistent AF patients compared with paroxysmal AF patients (25).
Several investigations have revealed that miR-126 is highly expressed in vascular tissues, including the heart, lungs, and liver. This miRNA is also expressed in hematopoietic and endothelial cells (30, 31). It has been shown that the expression of miR-126 levels may result in the activation of the vascular endothelial growth factor signaling pathway in the endothelium (32). Additionally, endothelial cell migration was disrupted during the processes of vascular growth, development, and organization after knocking down miR-126 expression in animal models. These processes are closely related to the development of AF and HF (33). Thus, miR-126 plays an important role in regulating the activation of the vascular endothelial growth factor pathway. Abnormal expression levels of miR-126 may cause impaired angiogenesis, leading to an elevated risk of AF and HF (34).
In this study, we observed that despite matching all risk factors between the patient and control groups, fasting serum glucose levels in patients with isolated atrial fibrillation were significantly higher than in control subjects. It has been established that AF patients with Diabetes Mellitus (DM) are susceptible to cardiovascular events and mortality (35). Such patients have a poor prognosis, decreased quality of life, and a higher risk of mortality and hospitalization compared to those without diabetes (36).
Vascular complications in DM patients are caused by endothelial dysfunction. Low levels of miR-126 have been observed in apoptotic bodies and microvesicles in diabetic individuals. Treatment of endothelial-derived EVs with high glucose and a small amount of miR-126 leads to the loss of the regenerative capacity of ECs (29). Therefore, hyperglycemia may have a role in defective angiogenesis and endothelial remodeling in AF patients with DM by contributing to endothelial dysfunction and lowering miR-126 expression in the vascular endothelium.
miR-126 suppresses inflammation in endothelial cells under hyperglycemic conditions. miR-126, expressed by endothelial cells, plays a crucial role in developmental angiogenesis in vivo. The association of miR-126 with vascular disease, inflammation, and angiogenesis has been reported (37). Recent studies have shown that miR-126 is involved in vascular inflammation (31, 38). miR-126 suppresses inflammation in endothelial cells incubated with high glucose by modulating HMGB1 expression. This suggests that miR-126 may be a useful molecular target for managing CVD, atherosclerosis, and AF in individuals with diabetes (39). These studies can explain the high levels of glucose and miR-126 in patients with lone AF in our study.
The cardioprotective protein SIRT1 is one of the predicted targets of miR-199a, and its expression level was increased in the heart tissue of individuals with AF and also in the serum of patients with chronic CAD (40, 41). Overexpression of SIRT1 is hypothesized to be a compensatory mechanism to prevent the oxidative stress process that contributes to the pathogenesis of AF and CAD (40, 42). In a recent transcriptomic study, miR-199a was designated as an up-regulated microRNA in patients with paroxysmal AF after cardiac surgery (43). It seems that miR-199a plays an important role in other types of AF. In our study, due to the absence of any risk factors such as CAD, hypertension, diabetes, or heart diseases and abnormalities, this miR did not show a significant difference between patients and controls. Therefore, it could be proposed as a marker for the differential diagnosis of AF from other types.
5.1. Conclusions
Our results strongly indicate that the decreased expression levels of miR-126 are associated with lone AF and that its reduced expression level could serve as an excellent diagnostic marker. This change in expression may be one of the mediators that accelerate the development of pathophysiology and complications in lone AF.