1. Background
Prolactinoma, despite being benign, is accompanied by many severe clinical manifestations including amenorrhea, galactorrhea, and dysgenesis in women and infertility and sexual dysfunction in men (1). Its frequency in women is higher with the ratio of 10:1 and at around the age of 20 - 50 years (2). It is the most frequent type of pituitary adenomas, which accounts for hypersecretion endocrinopathy (3). The pituitary is responsible for many regulatory functions in the human body including growth, metabolism, and reproduction (4). Evaluating molecular pathogenesis could enhance the clarification of disease mechanisms and thus targeting efficacious therapeutic agents (5, 6). By emerging high throughput studies, more knowledge about prolactinoma has been gained and a number of candidate biomarkers for clinical approaches have been identified. These agents can be essential for prediction, prevention, early-stage diagnosis, and treatment goals. Expression profiling is one of the ways of introducing some important elements of that specific disease (7, 8). Protein interaction analysis, on the other hand, can provide further insight into understanding biomarkers’ prominent roles and give more credit to their contributions (9). In other words, it is possible to assign the most vital ones by considering interaction characteristics through screening the interactome profile. It can show which agents are more important regarding the roles in a network constitution (10). These fundamentals are known as key participants in the interaction system. Any alteration in these elements may promote the differential interaction profile, which produces altered phenotype (11). Sometimes, these phenotypes could be a manifestation of a particular type of disease. Detecting new candidate genes for various diseases via PPI network analysis can be a useful medical tool (12). Therefore, we investigated new possible molecular markers correlated with Prolactinoma in terms of protein mapping that could be applicable for medical management of this disease. For this purpose, the interaction pattern of human Prolactinoma samples was selected and derived from an expression profiling study entitled “Genomic characterization of human and rat prolactinomas” (8).
2. Methods
The seed genes for the protein interaction network were from a microarray web-available data reported in Tong et al. study in 2012 (8) with the following characteristics: the platform of GPL8300, Dataset = GDS4859, Series Accession: GSE36314, and ID: 200036314. Human samples of three controls and four Prolactinoma were investigated and consequently top ranked 250 genes with fold change (FC) ≥ 2 and p value ≤ 0.05 were indicated through GEO2R, the online engine of Gene Expression Omnibus (GEO) screening (13). Similarly, the GEO2R provides a specific R formula for conducting the analysis in the R Studio environment using GEO query and limma R packages from the Bioconductor project (14). Genes with a differential pattern between healthy and Prolactinoma cases were further evaluated, and those with a gene name were used as seed genes for interaction network analysis. The network of the seeds and their neighbors was constructed by Cytoscape v 3.6.0 and its plug-in String dB (15, 16). Following the network restriction, Network Analyzer was used for centrality examination by considering specific parameters (17) including degree and betweenness of centralities to detect the potential elements of the network integrity. It is known that the removal of these nodes could pertubate the map organization and consequently, any abnormal phenotype may be accompanied by it (17). Nodes with the highest amount of the designated centrality parameters (degree and betweenness of centrality) are known as hub-bottlenecks. In fact, hub nodes are those with the highest value of degree and simultaneously, nodes with the highest amount of betweenness are considered as bottlenecks (18). The differentially expressed genes and hub-bottlenecks were chosen for more study namely, gene annotation. The ontology analysis assists in the better understanding of the important biological features of the designated agents. Here, via the application of Clue GO, the highlighted biological processes were assigned for our genes. The statistical criteria for this procedure are described in the legends of related tables. Bonferroni step-down was the used test for p-value correction. In addition, two-sided (enrichment/depletion) tests based on hypergeometric distribution for terms and groups were selected (19).
3. Results
Human prolactinoma expression profile (available in the GEO database) was used for interaction analysis in this study. First, via GEO2R, the samples were defined as groups of three human normal pituitaries and four human prolactinoma samples. Then, the value distribution of groups was determined in a way that boxplot assessed whether the expression values of control and Prolactinoma tissue samples were comparable in terms of expression pattern via cross-comparison (Figure 1)
Boxplot indication of median-centered samples of control and Prolactinoma. The blue boxes are the three control samples and the pink ones are Prolactinoma samples. The x-axis and y-axis indicate the range of expression values and biological replications for control and Prolactinoma, respectively. The comparison shows that the values are median-centered and consequently, the groups are comparable regarding the expression values.
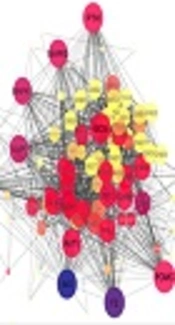
The next step is to make the comparison via GEO2R and detect differentially expressed genes across experimental conditions (control and human Prolactinoma tissue samples). GEO2R provides R script that was applied in R studio for the statistical analysis. Considering the fold change ≥ 2, up-regulated and down-regulated elements are presented in Tables 1 and 2, respectively. Among 59 genes, 13 are repeated corresponding to verities of the genes. A network of 46 identified genes by String dB plus 50 neighbor nodes is constructed considering the confidence score cut-off of 0.5. All the queried genes were retrieved as a complex interacting network. The network consists of overall 96 nodes and 1262 edges including a main connected component and four isolated nodes (three isolated query nodes; N4BP2L1, DLEU1, DLEU, and PLXNC1from the added nodes) (Figure 2). The hub-bottlenecks of the main connected component of the network are identified and listed in Table 3.
| Row | Gene Name | Gene Title |
|---|---|---|
| 1 | B2M | Beta-2-microglobulin |
| 2 | IGSF1 | Immunoglobulin superfamily member 1 |
| 3 | SV2C | Synaptic vesicle glycoprotein 2C |
| 4 | B2M | Beta-2-microglobulin |
| 5 | GNAS | GNAS complex locus |
| Row | Gene Name | Gene Title |
|---|---|---|
| 1 | GH1 | Growth hormone 1 |
| 2 | POMC | Proopiomelanocortin |
| 3 | TSHB | Thyroid stimulating hormone beta |
| 4 | GH2 | Growth hormone 2 |
| 5 | RBP4 | Retinol binding protein 4 |
| 6 | DLK1 | Delta-like non-canonical Notch ligand 1 |
| 7 | GH2 | Growth hormone 2 |
| 8 | IGFBP5 | Insulin-like growth factor binding protein 5 |
| 9 | GH2 | Growth hormone 2 |
| 10 | FSHB | Follicle stimulating hormone beta subunit |
| 11 | IGFBP5 | Insulin-like growth factor binding protein 5 |
| 12 | CRYAB | Crystallin alpha B |
| 13 | SAT1 | Spermidine/spermine N1-acetyltransferase 1 |
| 14 | CEBPD | CCAAT/enhancer binding protein delta |
| 15 | HBB | Hemoglobin subunit beta |
| 16 | GH1 | Growth hormone 1 |
| 17 | CSHL1 | Chorionic somatomammotropin hormone-like 1 |
| 18 | CSH2 | Chorionic somatomammotropin hormone 2 |
| 19 | CSH1 | Chorionic somatomammotropin hormone 1 |
| 20 | PTN | Pleiotrophin |
| 21 | IGFBP3 | Insulin-like growth factor binding protein 3 |
| 22 | NEFM | Neurofilament, medium polypeptide |
| 23 | CGB2 | Chorionic gonadotropin beta subunit 2 |
| 24 | CGB1 | Chorionic gonadotropin beta subunit 1 |
| 25 | CGB8 | Chorionic gonadotropin beta subunit 8 |
| 26 | CGB7 | Chorionic gonadotropin beta subunit 7 |
| 27 | CGB5 | Chorionic gonadotropin beta subunit 5 |
| 28 | CGB3 | Chorionic gonadotropin beta subunit 3 |
| 29 | N4BP2L1 | NEDD4 binding protein 2 like 1 |
| 30 | SRGN | Serglycin |
| 31 | CDH1 | Cadherin 1 |
| 32 | PTPN13 | Protein tyrosine phosphatase, non-receptor type 13 |
| 33 | WFDC2 | WAP four-disulfide core domain 2 |
| 34 | TGFBR3 | Transforming growth factor beta receptor 3 |
| 35 | SV2B | Ssynaptic vesicle glycoprotein 2B |
| 36 | ADD3 | Adducin 3 |
| 37 | TBL1X | Transducin (beta)-like 1X-linked |
| 38 | ALDH2 | Aldehyde dehydrogenase 2 family (mitochondrial) |
| 39 | NR3C1 | Nuclear receptor subfamily 3 group C member 1 |
| 40 | GSTP1 | Glutathione S-transferase pi 1 |
| 41 | ITPR1 | Inositol 1,4,5-trisphosphate receptor type 1 |
| 42 | GHRHR | Growth hormone releasing hormone receptor |
| 43 | CGB2 | Chorionic gonadotropin beta subunit 2 |
| 44 | CGB1 | Chorionic gonadotropin beta subunit 1 |
| 45 | CGB8 | Chorionic gonadotropin beta subunit 8 |
| 46 | CGB7 | Chorionic gonadotropin beta subunit 7 |
| 47 | CGB5 | Chorionic gonadotropin beta subunit 5 |
| 48 | CGB3 | Chorionic gonadotropin beta subunit 3 |
| 49 | ACTG1 | Actin gamma 1 |
| 50 | ACTB | Actin beta |
| 51 | ITPR1 | Inositol 1,4,5-trisphosphate receptor type 1 |
| 52 | DLEU1 | Deleted in lymphocytic leukemia 1 |
| 53 | GSTP1 | Glutathione S-transferase pi 1 |
| 54 | PLXNC1 | Plexin C1 |
| Row | Display Name | Description | Degree | BC |
|---|---|---|---|---|
| 1 | POMC | Proopiomelanocortin | 57 | 0.02 |
| 2 | F2 | Coagulation factor II (thrombin) | 54 | 0.07 |
| 3 | GCG | Glucagon | 54 | 0.02 |
| 4 | PTH | Parathyroid hormone | 53 | 0.02 |
| 5 | INS | Insulin | 52 | 0.08 |
| 6 | AKT1 | V-akt murine thymoma viral oncogene homolog 1 | 51 | 0.02 |
| 7 | MAPK3 | Mitogen-activated protein kinase 3 | 50 | 0.03 |
| 8 | AVP | Arginine vasopressin | 49 | 0.04 |
| 9 | MAPK1 | Mitogen-activated protein kinase 1 | 48 | 0.04 |
| 10 | GHRH | Growth hormone releasing hormone | 45 | 0.02 |
| 11 | IGF1 | Insulin-like growth factor 1 (somatomedin C) | 44 | 0.02 |
| 12 | CGA | Glycoprotein hormones, alpha polypeptide | 42 | 0.04 |
| 13 | ALB | Albumin | 42 | 0.02 |
| 14 | FSHR | Follicle stimulating hormone receptor | 42 | 0.02 |
| 15 | BRD2 | Bromodomain containing 2 | 41 | 0.05 |
aThe top 20% of the nodes (18 genes) based on degree value were selected as hub nodes and in a similar manner, the bottleneck nodes were identified. Common genes of the hub and bottleneck nodes (about 83%) were selected as hub-bottleneck genes. The genes are ordered by their degree value
Following the centrality analysis, ClueGO performed gene ontology of the 46 differentially expressed genes and the hub-bottleneck nodes based on biological processes (Tables 4 and 5).
| R | Term Group | Terms / Total Terms, % |
|---|---|---|
| 1 | Positive regulation of insulin-like growth factor receptor signaling pathway | 50 |
| 2 | Response to growth hormone | 30 |
| 3 | Platelet aggregation | 13 |
| 4 | Peptide hormone processing | 7 |
aStatistical criteria are as follows: Genes per term: 3, genes per term percent: 4, Kappa score: 0.5, Corrected P-value < 0.05, Grouping level: Min = 2, Max = 8 for term grouping.
| R | Term Group | Terms / Total Terms, % |
|---|---|---|
| 1 | Regulation of glycogen metabolic process | 40 |
| 2 | Regulation of phosphatidylinositol 3-kinase signaling | 30 |
| 3 | Positive regulation of nucleotide metabolic process | 15 |
| 4 | Phosphatidylinositol 3-kinase signaling | 10 |
| 5 | Killing of cell in other organisms | 5 |
aStatistical criteria are as follows: Genes per term: 3, genes per term percent: 4, Kappa score: 0.5, Corrected P-value < 0.05, Grouping level: Min = 2, Max = 8 for term grouping.
4. Discussion
Prolactinoma, while not malignant, can exert vast adverse effects on the human body (20). Molecular studies can be beneficial for understanding the disease mechanisms of onset and development and possibly reduction of the complicated side effects by identification of novel biomarkers. Protein-protein interaction network analysis is one of which providing essential information related to novel elements in a systematic interaction (12). Here, the interaction concept is based on the gene expression profile of a previous study conducted by Tong et al. in 2012 (8). First, seven samples consisting of three controls and four prolactinoma biological replications were compared in terms of expression quality in Figure 1. As can be inferred, the data are normalized and appropriate for proceeding the data analysis. The groups were then followed for the expression comparison and as shown in Tables 1 and 2, there are some genes considering the designated statistical criteria assigned as up-regulated and down-regulated, respectively. This analysis shows that most of the differentially expressed genes (92%) are down-regulated. In addition, it can be inferred that the differentially expressed genes are mostly involved in regulatory functions including growth, metabolic, and reproductive matters, which are the main responsibilities of the pituitary gland (20). Therefore, it is clear that these functions may be influenced in prolactinoma leading to many abnormal features. The next step was to examine differentially expressed genes as an interactome scale, as presented in Figure 2. In this network, there are some genes with additional properties known as central genes. These central elements are listed in table 3 and among them, only POMC is from differentially expressed genes. There is evidence that it is corresponding to the increased level of POMC in pituitary adenoma relative to the normal pituitary. The effect of POMC on alpha-melanocyte stimulating hormone leads to the regulation of melanin production (21). As indicated in our study, there is a chance that some of the central nodes are not among the query ones (22). As shown in table 3, the most central genes are identified among the added genes, implying the ability of network method to introduce some new therapeutic candidates related to differential genes playing a major role in interaction system. The significant roles of such highlighted genes on the integrity of the network are corresponding to their possible high impact on the pathology of the disease. For example, the presence of coagulation factor 2 as a central gene may indicate the blood coagulation process changes in Prolactinoma, in which the Hypercoagulable state was previously reported in Prolactinoma (23). Furthermore, other central genes are mostly metabolic and growth-related regulators, which directly or indirectly are related to the pituitary gland. Parathyroid hormone, insulin, glucagon, growth hormone-releasing hormone, insulin-like growth factor 1, and follicle stimulating hormone receptor, which comprise 40% of all central nodes, are mediated by the pituitary gland (24-27).
Clinical approaches indicate that impaired metabolic condition including serum glucose, cholesterol, and triglycerides occur in Prolactinoma patients (20). In this respect, some functional correlations between differential genes and central ones are present to play roles in Prolactinoma metabolic profile. For instance, insulin and insulin-like growth factor (IGF1) as one of the highly ranked central nodes in our network constitution are related to insulin-like growth factor binding proteins (IGFBPs), which belong to the down-regulated genes category. In this regard, insulin is reported to be responsible for inhibiting IGFBP-1 and IGFBP-2 (28). On the other hand, as mentioned earlier, one of the altered metabolites is serum fasting glucose of these patients (20), which could justify the linkage and importance of our identified central genes namely IGF1 and INS in Prolactinoma metabolic changes. This network indicates that how one part of the focused interactions could be responsible for Prolactinoma risk. Consequently, our screening method discriminated data effectively to provide a better molecular aspect of prolactinoma.
To achieve a better resolution of these prominent genes and the differentially expressed genes, their associated biological processes were also examined. As shown in tables 4 and 5, metabolic and growth regulation are the most highlighted processes of the hub-bottleneck and differentially expressed genes and possibly the outermost disrupted ones. The main features of biological processes related to differentially expressed genes are characterized as growth courses (Table 4) while based on the content of Table 5, the identified central nodes are mostly involved in metabolic pathways. Both metabolic and growth processes change grossly in cancer, which were also changed based on our findings in Prolactinoma. While there are many differential genes corresponding to prolactinoma pathogenesis, here we highlighted the crucial ones in terms of interaction pattern. As indicated above, our finding is consistent with previous investigations into prolactinoma. It is suggested focusing more on our introduced panel of central genes to get a better notion of their feasible participation in Prolactinoma pathogenesis.
4.1. Conclusion
The study declares that vast metabolic processes and growth functions are modified in Prolactinoma. The central genes that were introduced as a candidate biomarker panel in our study may be useful for clinical approaches including patient follow-up, diagnosing, and drug targeting for Prolactinoma. In this respect, conducting experimental assessments as the validation test is appreciated to examine their potential application in clinical fields.
 (1).jpg)

