1. Background
Klebsiella pneumoniae is one of the most important causative agents both in the form of community and hospital-acquired infections such as urinary tract infections, septicemia, pneumonia, and meningitis (1). Multidrug resistance has clinically developed anxiety, mainly for the elderly and immunocompromised patients. Also, infants with immature body physiology have the least ability against infection from multi drug-resistant (MDR) pathogens (2).
Antimicrobial resistance can happen due to a variety of mechanisms, including drug target alteration, enzymatic inactivation or modification of drug, and the prevention of drug penetration or accumulation. Efflux pumps are essential in drug resistance among Gram-negative bacteria as they actively pump antibiotics out of the cell and cause the accumulation of drugs inside the bacterial cell to decrease. Efflux pumps are a major concern as they can generate a concurrent resistance to several drugs (3).
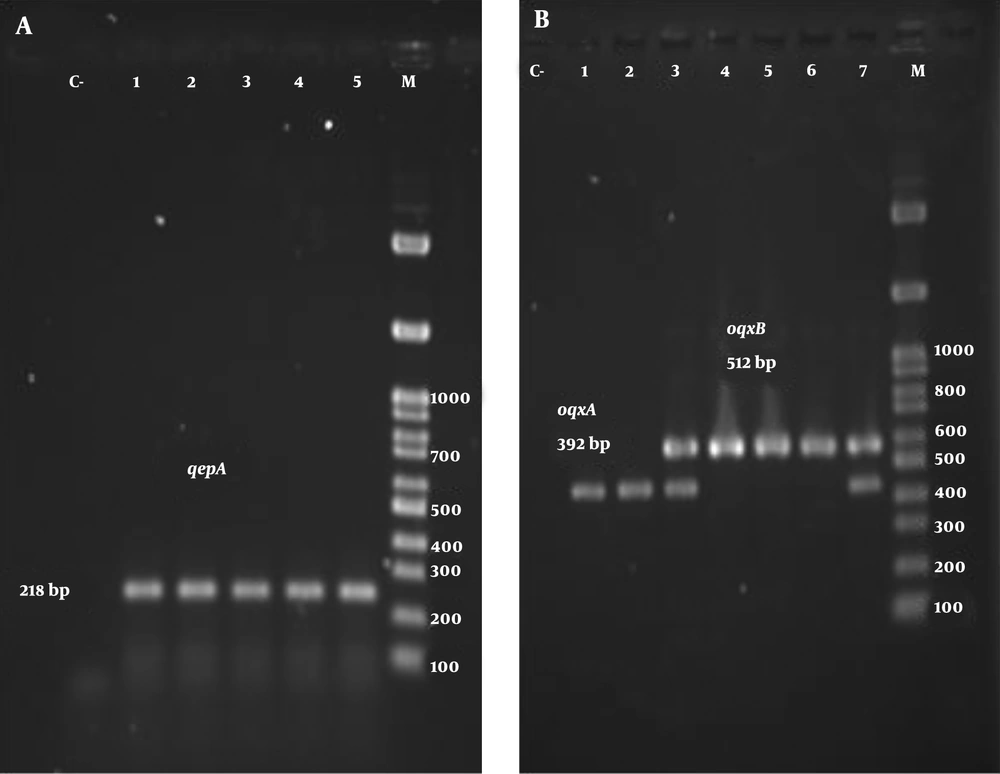
Among the efflux pumps, the resistance-nodulation-division (RND) family, including OqxAB and AcrAB, are important in creating antibiotic resistance in Gram-negative bacteria, especially K. pneumoniae. The OqxAB pump consists of two main domains: OqxA, a periplasmic part, and OqxB, a transmembrane protein, whose genes are located on both the chromosome and the plasmid (4). The OqxAB multidrug efflux pump is responsible for developing the reduced susceptibility or resistance to fluoroquinolones, such as flumequine, ciprofloxacin, and norfloxacin (2). In addition to quinolones, they confer resistance to other antibiotics, disinfectants, detergents, and antiseptics (5). A high prevalence of chromosomal or plasmid-mediated OqxAB has been reported in K. pneumonia, indicating a potential pool of this resistance determinant (2, 6, 7).
Another quinolone-specific efflux pump is QepA, which belongs to the major facilitator subfamily (MSF). QepA is a 14-transmembrane-segment MFS transporter encoded by the qepA gene and causes a decreased accumulation of norfloxacin (3). Minimum inhibitory concentrations (MICs) of ciprofloxacin, enrofloxacin, and norfloxacin were 32- to 64-fold higher in QepA-expressing test strains compared to host strains (8).
2. Objectives
Given the importance of efflux pumps in creating drug resistance, this study aimed to detect OqxAB and Qep efflux pumps in clinical isolates of K. pneumoniae. The antibiotic resistance profile of the isolates and, phenotypic and genotypic association of resistance to antibiotics were also investigated.
3. Methods
3.1. Clinical Samples and Identification of Bacteria
In this cross-sectional study, directed from March 2018 to April 2019, 100 non-repetitive urine isolates of K. pneumoniae were obtained from Milad Hospital in Tehran, Iran. Each urine sample was cultured on the Mac Conkey agar, and blood agar (Merck, Germany) followed by incubation at 37°C for 18 - 24 h. The isolates were then identified as K. pneumoniae by common microbiological and biochemical tests, including Gram-staining, catalase, oxidase, MR-VP, urease production, citrate utilization, and motility (9).
3.2. Antimicrobial Susceptibility Testing
Antibiotic susceptibility test was conducted via disc diffusion assay agreeing with Clinical and Laboratory Standards Institute (CLSI) instruction with the following antibiotics: ciprofloxacin (CIP: 5 µg), amikacin (AK: 30 μg), trimethoprim-sulfamethoxazole (TS, 2.5 μg), cefotaxime (CTX: 30 μg), nalidixic acid (NA: 10 μg), imipenem (IPM: 10 μg), gentamicin (GEN: 10 μg), ceftazidime (CAZ: 30 μg), ceftriaxone(CRO: 30 μg), meropenem (MER: 10μg), and levofloxacin (LEV: 5 μg) ( Mast, UK). In short, a bacterial suspension (equivalent to 0.5 McFarland) was prepared from overnight cultures and spread onto Mueller-Hinton agar (Oxoid, UK) plates. After the incubation at 37°C for 24 h, the diameter of inhibition zones around the discs was measured, and the results were expressed as resistant, susceptible, and intermediate. The standard strain of Escherichia coli ATCC 25922 was applied as control (10).
3.3. Detection of oqxA, oqxB, and qep Efflux Genes
Total genomic DNA was acquired via a DNA extraction kit (CinnaGen, Iran) based on the instructions of the kit manufacturer. The primer sequences used in this study are listed in Table 1. PCR was conducted in a volume of 25 µL PCR reaction mixture including 1 µL DNA template, 1 µL of each primer, 12.5 µL of PCR master mix (Amplicon Co., Denmark), and 10.5 µL of ddH2O. The PCR reaction was conducted with the following PCR procedure: initial denaturation at 95°C for 1 min, denaturation at 95°C for 45 s, annealing at 60°C for 45 s, extension at 72°C for 1 min (35 cycles), and the final extension at 72°C for 5 min. In the end, 10 µL of the amplicons were detected on 1% agarose gel electrophoresis.
The Sequences of Used Primers
3.4. Statistical Analysis
The association between the resistance to antibiotics and the presence of efflux genes was analyzed by SPSS version 20 using the chi-square and Spearman correlation coefficient tests, and a P value less than 0.05 was considered significant.
4. Results
4.1. Study Population
The mean age of the population studied was 52 ± 1.8 years, ranging from 13 to 81 years. The isolates were collected from patients with different age groups: 10 - 25 years no.; 18, 26 - 40 years no.; 29, 41 - 55 years no.; 36, 56 - 60 years no.; 10 and 60 - 81 years no.; 7. Seventy (70%) patients were female, and thirty (30%) patients were male.
4.2. Antibiotic Susceptibility Profiles
Table 2 shows the antibiotic susceptibility testing results. Accordingly, the highest resistance rate was observed against trimethoprim-sulfamethoxazole (72%), amikacin (70%), levofloxacin (68%), and the lowest resistance rate was observed against imipenem (10%). The frequency of MDR strains was 31%.
| Antimicrobial agents | Susceptible | Intermediate | Resistant |
|---|---|---|---|
| Gentamicin | 41 (41) | 3 (3) | 56 (56) |
| Amikacin | 30 (30) | 0 | 70 (70) |
| Imipenem | 78 (78) | 12 (12) | 10 (10) |
| Meropenem | 56 (56) | 10 (10) | 34 (34) |
| Ceftazidime | 38 (38) | 6 (6) | 56 (56) |
| Ciprofloxacin | 36 (39) | 15 (15) | 49 (49) |
| Levofloxacin | 27 (27) | 5 (5) | 68 (68) |
| Ceftriaxone | 39 (39) | 10 (10) | 51 (51) |
| Nalidixic acid | 62 (62) | 12 (12) | 26 (26) |
| cefotaxime | 54 (54) | 8 (8) | 38 (38) |
| trimethoprim-sulfamethoxazole | 26 (26) | 2 (2) | 72 (72) |
Antibiotic Susceptibility Profile of K. pneumoniae Isolatesa
4.3. Molecular Distribution of Resistance Efflux Pump Encoded Genes
The results of the PCR test for oqxA and oqxB genes showed that 57% and 56% of strains were the carriers of these genes, respectively. In addition, 52% of isolates carried both genes simultaneously. The results of the molecular test also showed that 21 isolates were positive for the qepA gene on the electrophoresis test, and 15 isolates harbored all three genes. Figure 1 depicts the electrophoretic pattern of studied genes.
Table 3 demonstrates the association between antibiotic resistance and the presence of efflux genes. As shown, resistance to fluoroquinolones and beta-lactams was associated with the presence of oqxA/oqxB genes. qepA gene was also associated with resistance to levofloxacin, imipenem, meropenem, and ceftriaxone (P < 0.05).
| Efflux Genes | Ciprofloxacin | Levofloxacin | Imipenem | Meropenem | Ceftriaxone | Ceftazidime |
|---|---|---|---|---|---|---|
| oqxA/oqxB | 0.001 | 0.001 | 0.025 | 0.001 | 0.001 | 0.001 |
| qepA | 0.243 | 0.036 | 0.032 | 0.025 | 0.001 | 0.07 |
Significant Relationships Between the Efflux Genes and Antibiotic Resistance
5. Discussion
K. pneumoniae is one of the most important causative agents of community and hospital-acquired infections worldwide. Awareness of antimicrobial susceptibility and genetic determinants that contribute to resistance in this bacterium can help control infections and reduce mortality and morbidity. The high resistance rate was related to ciprofloxacin (49%), amikacin (70%), trimethoprim-sulfamethoxazole (72%), gentamicin (56%), ceftazidime (56%), and levofloxacin (68%). In contrast with our data, Amiri et al. (13), posited that resistance to cefotaxime, ceftriaxone, and ceftazidime was 59%, 65%, and 67%, respectively. This conflict may be related to the year of study. Resistance to some antibiotics has increased significantly over the years. For instance, in the present study, 70% of the isolates were resistant to amikacin, while in studies conducted in 2014, 26% (14) in 2015, 35% (15), and in 2019, 65% (16) of isolates were resistant to amikacin. In the present study, resistance to ciprofloxacin increased from 30% in 2013 (17), 33% in 2015 (15), and 43% in 2019 (18), to 49%. This increasing resistance to antibiotics indicates the indiscriminate and uncontrolled use of antibiotics, as well as the acquisition of numerous antibiotic resistance mechanisms in the K. pneumoniae. One of the most worrisome findings in this study was the resistance of K. pneumoniae strains to imipenem. Since carbapenems are used as the first-line drugs, the treatment of antimicrobial-resistant infections (19), resistance to imipenem can lead to serious problems in the treatment of K. pneumoniae infections.
The results of the PCR test for oqxA and oqxB genes showed that 57% and 56% of isolates were carriers of these genes, respectively. Also, 52% of isolates carried both genes simultaneously. The prevalence of oqxA and oqxB genes in the study conducted by Zomorrodi et al. (18) was 69.7% and 72.1%, respectively. Higher prevalence of oqxA and oqxB were obtained among K. pneumoniae strains isolated in Spain (76% and 75%, respectively) (7). In our study, 21% of K. pneumoniae isolates harbored the qepA gene, which was higher than those reported by Goudarzi et al. (20), (2%) and Heidary et al. (1), (4%). The genetic diversity of strains, geographical distance, type of samples, and the number of samples are among the main reasons for the discrepancy between the prevalence of efflux genes in the present study and other studies.
Numerous studies have shown that efflux pump systems are involved in the resistance to the fluoroquinolones (7, 21, 22). In this study, 83.7% of ciprofloxacin-resistant strains had the oqxA/oqxB efflux genes. The frequency of these genes was significantly higher in the ciprofloxacin-resistant isolates than ciprofloxacin sensitive isolates (P = 0.001). In addition, 77.8% of levofloxacin-resistant isolates harbored oqxA/oqxB, and a significant association was observed between levofloxacin resistance phenotype and the presence of oqxA/oqxB efflux genes. Levofloxacin resistance was also associated with the presence of the qepA efflux gene (P < 0.05); however, no association was observed between ciprofloxacin resistance and the presence of the qepA gene. Interestingly, the presence of oqxA/oqxB efflux genes was significantly associated with resistance to imipenem, meropenem, ceftriaxone, and ceftazidime, which has not been reported earlier. To date, there have been no reports regarding the association of the qepA efflux gene with beta-lactam resistance. Further studies are needed to investigate the role of these pumps in beta-lactam resistance in K. pneumoniae isolates.
5.1. Conclusions
The high frequency of efflux genes in this study is a serious concern, so we require a careful control of drug administration and identification of resistant isolates to control infection and prevent the spread of drug-resistant bacteria. We found a significant association between the presence of the efflux genes and resistance to fluoroquinolone and beta-lactam antibiotics, which signifies the need for novel treatment strategies to reduce the incidence of resistance.