1. Background
Listeria monocytogenes is a facultative, intracellular, gram-positive bacterium with the remarkable ability to adapt and grow in diverse environments (1). While symptoms can greatly vary, listeriosis predominantly affects high-risk individuals, such as the elderly, immunocompromised individuals, pregnant women, and their unborn children (2).
Listeria monocytogenes is transmitted through food, particularly when ready-to-eat products become contaminated during the manufacturing process. These bacteria have the unique ability to multiply even at refrigerator temperatures, posing a risk, especially when food is consumed without sufficient heating (3). Infections with L. monocytogenes during pregnancy account for 16 - 27% of all listeriosis cases. Despite antibiotic treatment, the invasive form of listeriosis carries a high mortality rate, particularly among individuals with compromised immune systems (4). Nanoparticles (NPs) are garnering significant attention as potential new antibacterial agents. They can have a variety of applications, including drug delivery, gas sensors (5), chemical and biological sensors, CO2 capture, and other related applications (6).
Chitosan nanoparticles (CNPs) are known for their biocompatibility, biodegradability, and relatively non-toxic nature, along with their cationic properties (7). They are widely recognized and utilized in various biological applications, including drug delivery, due to their acknowledged properties. The method most commonly used for the production of CNPs is known as ionic gelation (8). In this study, we utilized real-time polymerase chain reaction (RT-PCR) to examine the expression of the ftsL gene, which is involved in cell division, as well as the pflA gene, which encodes a fermentation enzyme.
2. Objectives
This study aimed to investigate how CNPs influence the expression of the pfLA and ftsL genes in L. monocytogenes strains.
3. Methods
3.1. Bacterial Strain
In this study, five strains of L. monocytogenes were isolated from vaginal samples of pregnant women collected by Kazemi Rad et al. Additionally, the standard strain of L. monocytogenes, ATCC 7644, was examined (9).
3.2. CNP Preparation
In this study, CNPs produced by Alebouyeh et al. (10) were utilized. Shells from green shrimp (Penaeus semisulcatus) were cleaned, thoroughly dried, and ground into powder using an electric mill. To extract chitin, 10 g of powdered shrimp was stirred in 150 mL of 4% hydrochloride for 30 minutes in a shaker incubator at a temperature of 25°C for 30 minutes. The solution was then centrifuged, and the powder was washed repeatedly with distilled water until it reached a neutral pH, as measured by a pH meter. Subsequently, the neutralized powder was thoroughly dried. The transformation of chitin into chitosan (CS) was achieved using a heated sodium hydroxide (NaOH) solution at a concentration of 40% to 50% (w/w).
The production of CNPs was initiated with the dissolution of 20 mg of CS in 40 mg of 1% acetic acid. The pH of the solution was then adjusted to 5.5 using a 5% molar NaOH solution. Next, a 25% solution of sodium tripolyphosphate (STPP) was prepared. Following this, 2 mL of the STPP solution was added dropwise to 12 mL of the CS solution at room temperature while stirring vigorously for an hour to ensure the complete formation of the NPs. The NPs were then formed by centrifuging the solution for approximately 45 minutes at 5000 rpm. Subsequently, they were dried and converted into a powder form. Next, 8.00 g of the powder was dissolved in 1 mL of distilled water. This solution was then filtered using a syringe filter and prepared for microbial testing (10).
The physicochemical properties of CNPs, including zeta potential, particle size, and polydispersity index (PDI), were determined using the dynamic light scattering method with a Zetasizer (Nano ZS, Malvern, UK).
3.3. Antimicrobial Activity
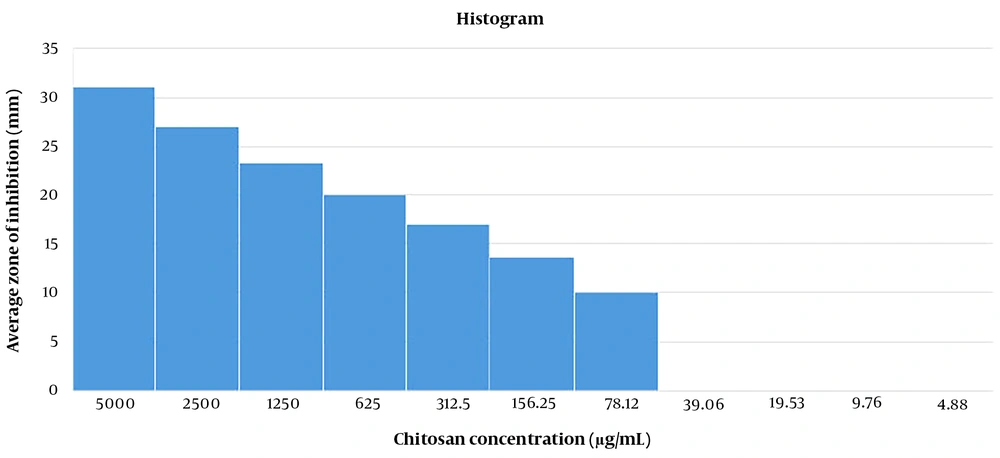
The antimicrobial activity of CNPs was evaluated using the well diffusion method. In this procedure, holes were made in a Müller-Hinton agar medium that had been previously cultured with L. monocytogenes strains. Subsequently, 20 mL of CNPs were added to these wells at various concentrations (range, 4.88 - 5000 μg/mL). The plates were then incubated at 37°C for 24 hours. The inhibition zone diameter was finally measured (9).
3.4. Minimum Inhibitory Concentration
The minimum inhibitory concentration (MIC) value was determined using the macrodilution test. This test was used to evaluate the sensitivity of L. monocytogenes strains to the CNPs. To calculate the MIC, a fresh bacterial solution was prepared, containing a concentration of 0.5 McFarland (equivalent to 1.5 × 108 cells/mL). Next, a Müller-Hinton broth was prepared with varying concentrations of NPs, ranging from 4.88 to 5000 μg/mL. This broth was then incubated for a period of 24 hours at a temperature of 37°C. Following this, a 100 μL aliquot of the microbial suspension was introduced into each tube. The MIC value was determined as the lowest concentration of the antimicrobial agent that completely inhibited visible bacterial growth. For molecular evaluations, the concentration right below the MIC, known as the sub-MIC, was calculated (11, 12).
3.5. Evaluation of pfLA and ftsL Gene Expression
The RT-PCR assay was used to compare the molecular alterations caused by CNP on the expression of the pfLA and ftsL genes in L. monocytogenes. Six different strains of L. monocytogenes were grown for 24 hours in tryptic soy broth with yeast extract (TSBYE) at 37°C in two experimental stages. Next, a suspension of each strain was prepared at a concentration of 0.5 McFarland. The impact of NPs on gene expression was examined based on the sub-MIC concentrations. Each experiment was conducted in triplicate. The accuracy of the test was assessed using an internal or positive control gene known as LDH. This gene, often referred to as a housekeeping gene, exhibits consistent expression across all samples and conditions (13, 14).
3.5.1. Primer Design
The direct (F) and reverse (R) primers were designed using the Gene Runner software (version 3) after the gene sequences were retrieved from the National Center for Biotechnology Information (NCBI) (Table 1).
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| LDH | ATGGCAAGCGGATTTGATGG | TGACATACGGAAACGTGCTG |
| pflA | GCTGTTGTGTTAGCCCACTT | CCACCGTTGATAGCACCAAG |
| ftsL | AGTGTGAGCAATGTCGCCTA | TGTAAATTTGCGCTTGTACGC |
3.5.2. RNA Extraction
Following the manufacturer's instructions, total RNA was extracted using the Trizol Reagent (Yekta Tajhiz Azma Kit, Iran). According to the manufacturer's instructions, a cDNA synthesis kit (Yekta Tajhiz Azma cDNA Synthesis Kit, Iran) was utilized to convert 5 µg of total RNA into cDNA. The initial amplification of pflA and ftsL was performed using primers. This process began with a 15-minute initial denaturation step at 95°C, followed by a denaturation phase for 15 - 30 seconds at 95°C, primer annealing at 60°C for 30 seconds, and primer extension at 72°C for 30 seconds. After these steps, the samples were stored at 4°C. The RT-PCR assay was performed using a StepOnePlus RT-PCR machine with the Ampliqon SYBR Green Kit (Pishgam Biotech Co., Iran). In three separate studies, reactions were run in triplicate. The RT-PCR findings were analyzed using the -2ΔΔCT technique after the expression data were normalized with LDH to account for any variations in the expression levels (15).
3.6. Statistical Analysis
SPSS version 26 was used to compile and analyze the data. A P-value less than 0.05 was considered statistically significant. Graphs were also drawn using GraphPad Prism 9 software.
4. Results
4.1. Physicochemical Properties of CNPs
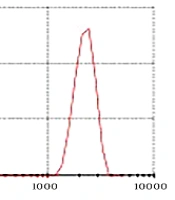
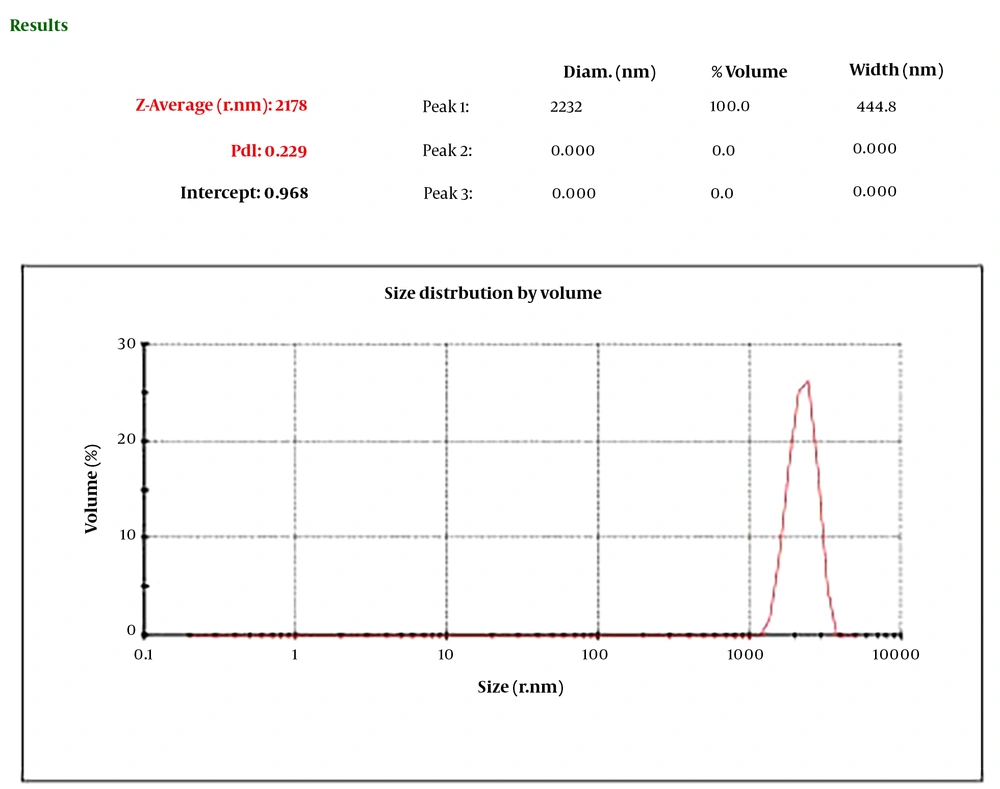
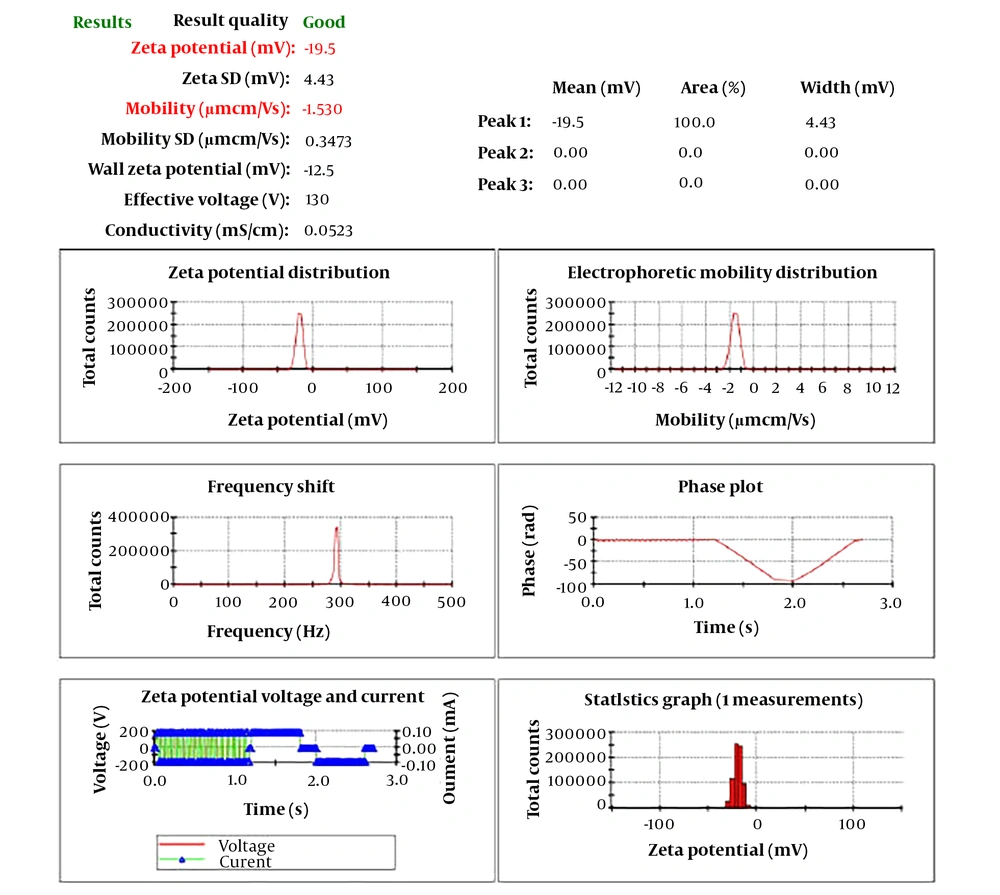
The CNPs had a diameter ranging from 22 to 80 nm and a purity of 99%. As depicted in Figure 1, the PDI was determined to be 0.229. Furthermore, according to Figure 2, the zeta potential of the CNPs was measured to be -19.5 mV.
4.2. Antimicrobial Activity
The results revealed that the antimicrobial effects of CNPs on L. monocytogenes strains were not significant at concentrations of 4.88 to 39.06 μg/mL. Therefore, these concentrations of CNPs do not influence the bacteria (P > 0.05) (Figure 3). Also, the antibacterial activities of CNPs were enhanced at higher CNP concentrations.
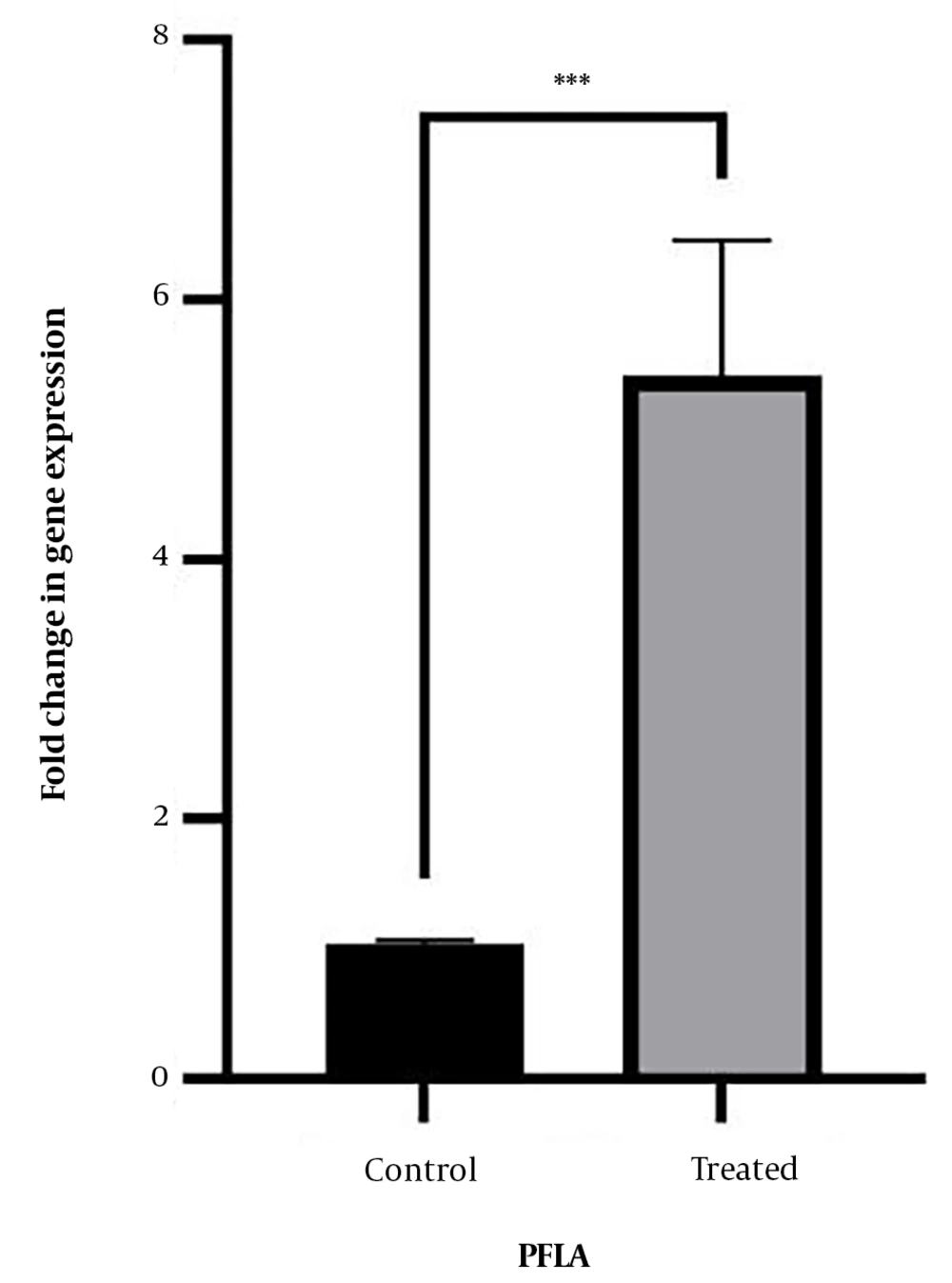
4.3. Results of pfLA Gene Expression in Standard and Clinical Strains
The impact of stress induced by the MIC concentration of CNPs (78.12 µg/mL) on the expression of the pfLA gene was investigated in L. monocytogenes strains. As depicted in Figure 4, the expression level of the pfLA gene increased in all strains of L. monocytogenes following exposure to the CNPs. The significance level (P-value) of the change in gene expression was found to be 0.0001.
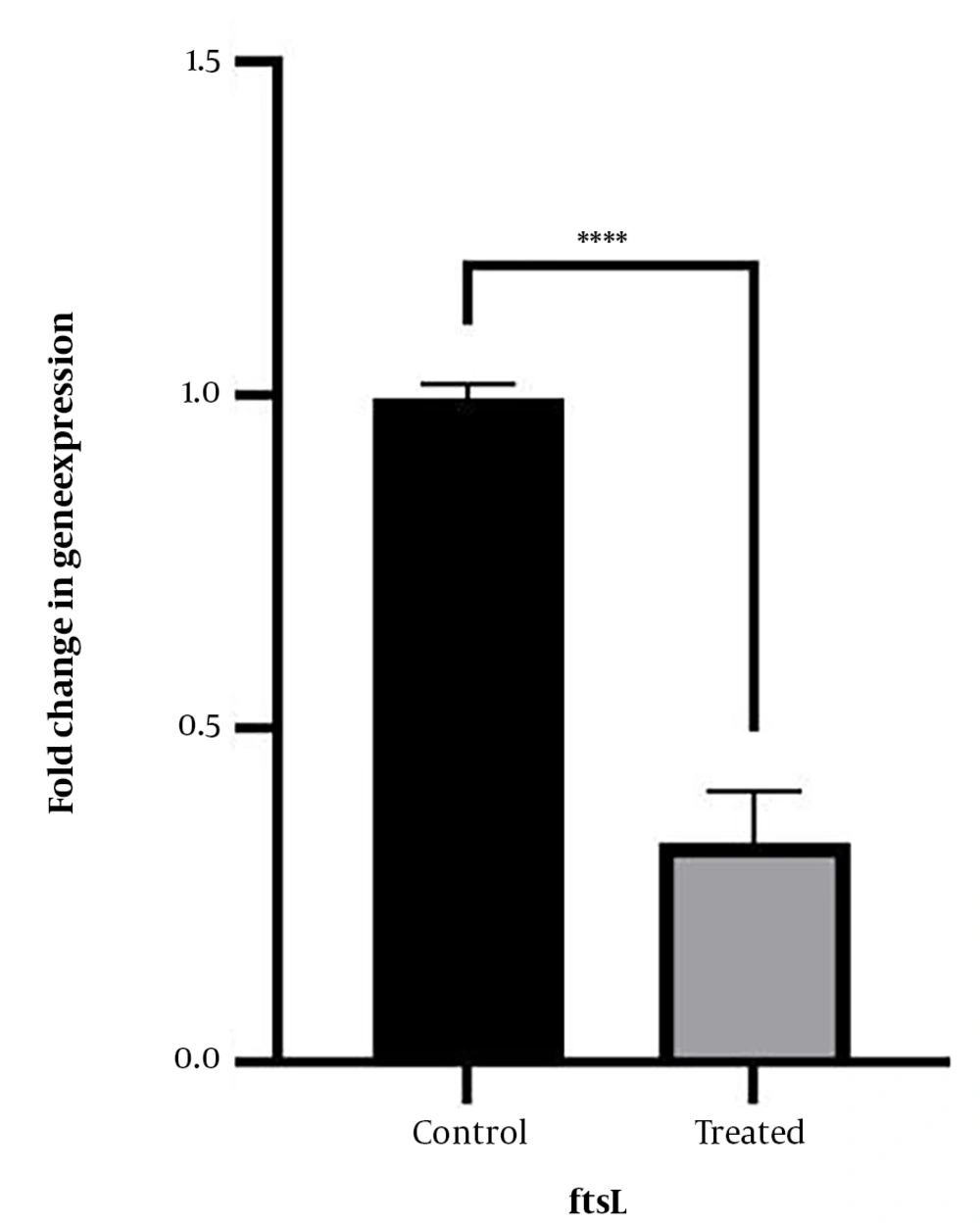
4.4. Results of ftsL Gene Expression in Standard and Clinical Strains
The impact of stress induced by the MIC of CNP (78.12 µg/mL) on the expression of the ftsL gene was examined in L. monocytogenes strains. As shown in Figure 5, the expression level of the ftsL gene decreased in all strains of L. monocytogenes following exposure to CNPs. The significance level (P-value) of this change in gene expression was found to be less than 0.0001.
5. Discussion
Listeria monocytogenes is a foodborne bacterium that causes listeriosis, a potentially fatal condition, particularly in individuals with compromised immune systems (16). In high-risk populations, the death rate of listeriosis may reach as high as 30% (17). According to a study by Mateus et al., pregnancy was a risk factor in 43% of patients with listeriosis infections across 20 nations. Pregnant women with undiagnosed listeriosis had a risk of miscarriage, early delivery, fetal mortality, or neonatal problems, such as sepsis, meningitis, and encephalitis (18).
Due to its biodegradable and non-toxic properties, CS has emerged as a popular choice with a broad spectrum of beneficial applications in fields such as biomedicine and agriculture. As nanotechnology has emerged as a new field, research has incorporated CNPs into various products to enhance their efficacy and biocompatibility. Chitin and CS hold the potential for a range of medicinal applications. Notably, they have the ability to modulate immune responses. The human immune system is capable of recognizing and degrading these natural polymers. As a response to these polymers, the immune system produces enzymes that can degrade chitin and chitosans, such as chitinases (19).
The low solubility of chitin is considered the most important factor limiting the use of this biopolymer. Despite this limitation, so far, many applications of chitin and its derivatives have been reported. When chitin is soluble in acidic aqueous solvents whose pH is < 6, it is known as CS. Owing to the presence of free amine groups along its polymer chain and its good solubility in weak acids, such as acetic acid, CS has secured a prominent position among polysaccharides (20).
Due to its polycationic nature at pH levels < 6, CS readily interacts with negatively charged substances. These include proteins, fatty acids, phospholipids, and anionic polysaccharides (21). Also, given its polycationic properties, CS destroys bacterial membranes and has antibacterial properties (22). Indeed, the antibacterial properties of CNPs can be attributed to the protonation of amino groups. These protonated amino groups can interact with the negatively charged cell surface, leading to the destruction of bacterial cells.
In 2017, Baradaran Ketabchi et al. used concentrations of 4.88 to 5000 μg/mL to evaluate the inhibitory activity of zinc oxide NPs against Staphylococcus aureus and Escherichia coli strains. In this research, we tried to investigate a wide range of CNP concentrations (11). According to a study by Mao et al., the antibacterial activities of CNPs were enhanced with higher particle concentrations. The lack of bacterial growth due to different concentrations of these NPs showed that the greatest inhibitory effect of CS on clinical strains and standard L. monocytogenes was related to higher NP concentrations; this implies that the inhibitory effect is enhanced by increasing the NP concentration (23).
In a 1972 study by Moellering et al. entitled "antibiotic synergy against 20 strains of L. monocytogenes", the effects of ampicillin, penicillin, streptomycin, and gentamicin were studied in vitro. The minimal bactericidal concentration (MBC) of ampicillin was higher than its MIC, while the MBC and MIC values of streptomycin and gentamicin were close. However, relatively higher concentrations were required to inhibit the growth of L. monocytogenes strains (24). In this regard, Derveaux et al. highlighted that reverse transcription quantitative PCR (RT-qPCR) is today the gold standard for sensitive, accurate, cost-effective, and rapid measurement of gene expression (25); therefore, we used this method to examine the expression of genes.
The process of bacterial cell division commences with the recruitment of the bacterial GTPase tubulin homolog, FtsZ, to the division plane. Following this, division proteins are summoned in a hierarchical and temporal manner. This sequence of events facilitates the synthesis of a peptidoglycan cell wall at the site of division. The bacterial GTPase tubulin homolog, FtsZ, and the Z-ring complex (including components such as ZipA and FtsA) are the initial components that are recruited to the division plane. Following this, there is a temporal delay before the recruitment of the peptidoglycan synthesis protein complex (including components such as FtsI and FtsW). A key set of proteins in this process is the FtsLBQ complex, most of which are essential in multiple bacteria (26).
According to a study by Diaz-Visurraga et al., exposure to antimicrobials, as well as unfavorable environmental conditions, can occasionally lead to stress in organisms. In our study, we observed a decrease in the expression of the ftsL gene, which plays a crucial role in cell division, following exposure to CNPs (27). Generally, PflA is a member of the pyruvate formate lyase protein family, which catalyzes the non-oxidative conversion of CoA and pyruvate into acetyl-CoA and formate. It supports microbial growth by facilitating the condensation of acetyl-CoA and formate. It is a key enzyme that enhances ATP yield during sugar fermentation in certain bacteria (28).
A study conducted by Raafat et al. in 2008 discovered that CS disrupts cellular energy metabolism. They found that several genes, which are typically expressed under hypoxic conditions, were upregulated, and their expression levels increased in the presence of CS. In the absence of oxygen and external electron acceptors, such as oxygen or nitrate, NAD+ is regenerated through fermentation or nitrate respiration, bypassing the respiratory chain. This process involves enzymes, such as pflB and pflA, which are involved in fermentation. These proteins exhibit the highest transcript levels under these conditions (14). In the present study, following exposure to CNPs, the expression of the pflA gene was observed to increase in both clinical and standard strains.
5.1. Conclusions
The current findings suggest that CNPs significantly contribute to the prevention of infections and complications associated with listeriosis. Furthermore, they appear to influence the expression of the pfLA and ftsL genes in L. monocytogenes.