1. Background
Staphylococcus aureus and Enterococcus faecium are the most significant bacterial pathogens, which have emerged over the past 30 years and are able to cause hospital acquired infections (1). These are the second most common organisms recovered from hospital associated urinary tract and wound infections and the third most common cause of hospital associated bacteraemia (2). These isolated strains are resistant to many antibiotic therapies and have become an important public health concern (3). Moreover, it is of particular concern if causative agents have multidrug resistance (MDR). Some organisms are resistant to all approved antibiotics. An alarming increase in resistance of bacteria that cause community acquired infections has also been documented, especially in the staphylococci and enterococci, which are prevalent causes of disease and mortality. In a recent study, 25% of bacterial infections were shown to be resistant to penicillin, and an additional 25% of cases were resistant to more than one antibiotic. Antimicrobial resistance among bacterial pathogens of hospital acquired infections is on the increase and the control of hospital acquired infections has become more challenging due to the wide spread of Staphylococcus aureus and Enterococcus faecium that are resistant to multiple antibiotics (4). Unless antibiotic resistance problems are detected as they emerge and actions are taken immediately, the society could be faced with previously treatable diseases that have become untreatable again, as in the days before antibiotics were developed.
2. Objectives
The present study aimed to observe the biochemical characteristics of Staphylococcus aureus and Enterococcus faecium and to determine the antibiotic resistance profiles of these two isolates from clinical impact sources of urine specimens.
3. Materials and Methods
3.1. Sample Collection and Handling
Clinical impact source ofmid-stream urine specimens were collected from K. A. P. Vishwanatham Government Medical College, Tiruchirappalli, Tamil nadu, India. A total of 50 clinical specimens were collected from hospital admitted patients. All the specimens were processed within 24 hours of collection.
3.2. Identification of Isolates
Morphological and physiological analysis was carried out by various microbiologically significant examinations including Gram staining, motility assessment and various important biochemical analysis such as coagulase test and catalase test, which were used to identify the growth of the bacteria.
3.3. Growth Media
Various types of media such as blood agar media, mannitol salt agar and esculin bile salt agar were used to identify the cultural growth of gram positive isolates.
3.4. Disc Diffusion Method
According to the guidelines of the Clinical and Laboratory Standard Institute (5), multi-drug resistance was detected by using the disk diffusion test which was performed on Muller Hinton agar medium. The plates were incubated at 37°C for 24 hours. Any growth with less than 12 mm in diameter zone around the disk was considered indicative of drug resistance to the bacterial growth.
3.5. Multiple Antibiotic Resistance Index
Multiple antibiotics resistance index was calculated by using following formula:
MAR Index = Number of antibiotics to which the isolate was resistant/Total number of antibiotics tested.
4. Results
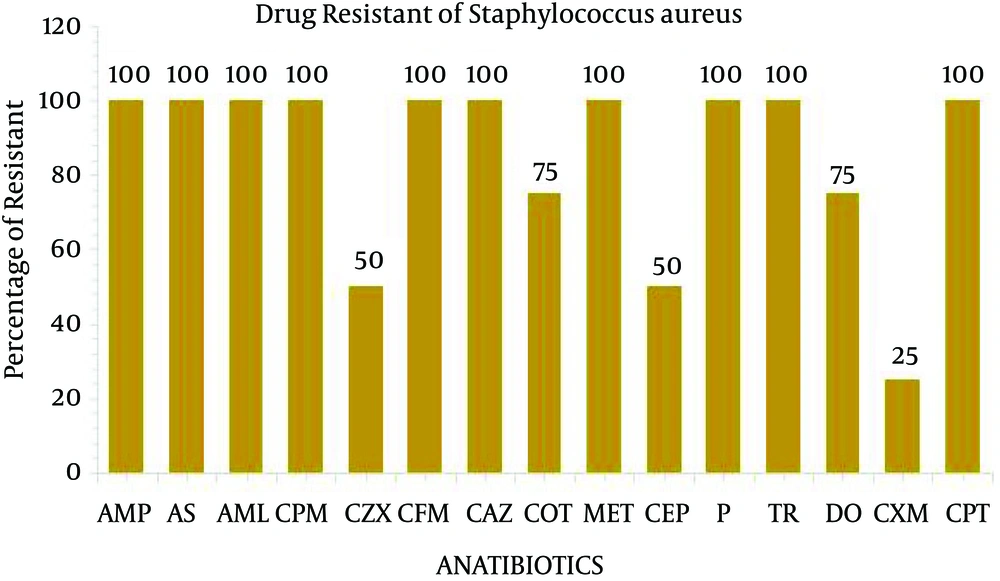
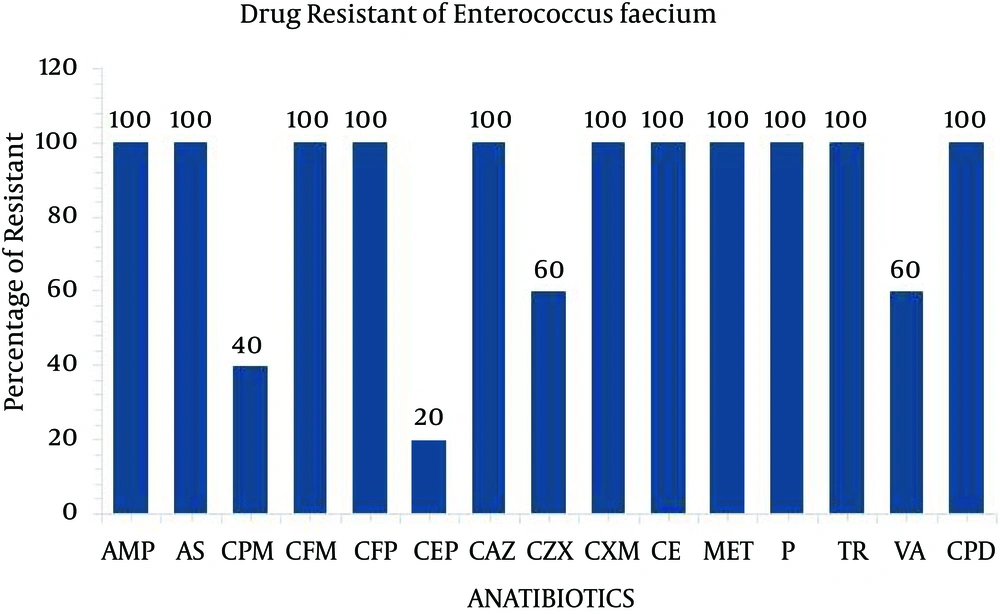
The present study performed the isolation, identification and antibiogram of multi-drug resistant gram-positive bacteria isolated from clinical impact source of urine specimens. According to Bergey’s manual of determinative bacteriology, the isolates were determined by using morphological and biochemical characterization. Among the 50 clinical urine specimens, Staphylococcus aureus was found in four clinical urine specimens and Enterococcus faecium was found in five specimens (Table 1). The isolate of Staphylococcus aureus was grown on Mannitol salt agar and exhibited a yellow color. On blood agar, Staphylococcus aureus exhibited beta hemolytic activity and the coagulase and catalase tests indicated positive growth. The isolate of Enterococcus faecium was grown in 6.5% NaCl at 10°C and nutrient agar medium at 45°C and black colored colonies were grown on bile esculin salt agar. The catalase and oxidase tests were considered as negative (Table 2). All four isolates of Staphylococcus aureus and five isolates of Enterococcus faecium were exposed to 35 different antibiotic discs, which were used to identify the drug resistance pattern (Tables 3 and 4). The four strains of Staphylococcus aureus were highly resistant to ampicillin, ampicillin sulbactam, amoxy/clav, cefixime, penicillin, methicillin, cefepime and ceftazidime (Figure 1). The five strains of Enterococcus faecium isolates were highly resistant to ampicillin, ampicillin sulbactam, methicillin, penicillin, cefotaxime, ceftazidime, cefpirome, cefuroxime, cefixime and trimethoprim and three strain of vancomycin (Figure 2). Multiple antibiotic resistance index was calculated as 0.428 for the two strains. These two strains were highly sensitive to various antibiotics such as amikacin, ciprofloxacin, ertapenem, gentamycin, levofloxacin, linezolid, ofloxacin, teicoplanin.
| Source of Urine Specimen for Staphylococcus aureus | Sample No. | Source of Urine Specimen for Enterococcus faecium | Sample No. | |
|---|---|---|---|---|
| 1 | Diabetes | 1 | Pregnancy | 2 |
| 2 | Pregnancy | 1 | Painful Urine | 1 |
| 3 | Pyuria | 2 | Blood in Urine | 1 |
| 4 | - | - | Transplanted Kidney | 1 |
| Total | 4 | 5 |
Types of Positive Urine Specimens Examined for the Isolation of Bacteria
| Test Name | Staphlococcus aureus | Enterococcus faecium | |
|---|---|---|---|
| 1 | Gram stain | Gram positive cocci | Gram positive cocci |
| 2 | Motility | Non motile | Non motile |
| 3 | Blood agar | Beta-haemolysis | No haemolysis |
| 4 | Coagulase test | Positive | Negative |
| 5 | Mannitol salt agar | Appearance of yellow colonies | No growth |
| 6 | Esculin bile salt agar | No growth | Appearance of block color colonies |
| 7 | 6.5% NaCl | No growth | Appearance of growth |
| 8 | Catalase | Positive | Negative |
Morphological and Biochemical Analysis of Isolates
| Antimicrobial Agent | Conc, mg | R, mm | I, mm | S, mm | |
|---|---|---|---|---|---|
| 1 | Amikacin | 30 | - | - | 4 |
| 2 | Ampicillin | 10 | 4 | - | - |
| 3 | Amoxy/clav | 20/10 | 4 | - | - |
| 4 | Ampicillin/sulbactam | 10/10 | 4 | - | - |
| 5 | Azithromycin | 15 | - | - | 4 |
| 6 | Cefepime | 30 | 4 | - | - |
| 7 | Cefepime/Tazobactam | 30/10 | 4 | - | - |
| 8 | Cefixime | 5 | 4 | - | - |
| 9 | Cefpirome | 30 | - | 1 | 3 |
| 10 | Cefpodoxime | 10 | - | 4 | - |
| 11 | Ceftazidime | 30 | 4 | - | - |
| 12 | Ceftazidime/Tazo | 30/10 | - | 4 | - |
| 13 | Ceftizoxime | 30 | 2 | 2 | - |
| 14 | Cefuroxime | 30 | 1 | 3 | - |
| 15 | Cephalothin | 30 | 2 | 2 | - |
| 16 | Cefotaxime | 10 | - | 4 | - |
| 17 | Ciprofloxacin | 5 | - | - | 4 |
| 18 | Co Trimoxazole | 25 | 3 | - | 1 |
| 19 | Doxycycline | 30 | 3 | - | 1 |
| 20 | Erythromycin | 15 | - | - | 4 |
| 21. | Ertapenem | 10 | - | - | 4 |
| 22 | Gentamycin | 10 | - | - | 4 |
| 23 | Imipenem | 10 | - | - | 4 |
| 24 | Levofloxacin | 5 | - | - | 4 |
| 25 | Linezolid | 30 | - | - | 4 |
| 26 | Methicillin | 5 | 4 | - | - |
| 27 | Meropenem | 10 | - | - | 4 |
| 28 | Moxifloxacin | 5 | - | - | 4 |
| 29 | Ofloxacin | 5 | - | - | 4 |
| 30 | Penicillin | 10 | 4 | - | - |
| 31 | Piper/Tazobactam | 100/10 | - | - | 4 |
| 32 | Teicoplanin | 30 | - | - | 4 |
| 33 | Tetracycline | 30 | - | - | 4 |
| 34 | Trimethoprim | 25 | 4 | - | - |
| 35 | Vancomycin | 30 | - | - | 4 |
Disc Diffusion Pattern of Staphylococcus aureus a
| Antimicrobial Agent | Conc, mg | R, mm | I, mm | S, mm | |
|---|---|---|---|---|---|
| 1 | Amikacin | 30 | - | - | 5 |
| 2 | Ampicillin | 10 | 5 | - | - |
| 3 | Amoxy/clav | 30 | - | 2 | 3 |
| 4 | Ampicillin/sulbactam | 10/10 | 5 | - | - |
| 5 | Azithromycin | 15 | - | - | 5 |
| 6 | Cefepime | 30 | 2 | 3 | - |
| 7 | Cefepime/Tazobactam | 30/10 | - | 2 | 3 |
| 8 | Cefixime | 5 | 5 | - | - |
| 9 | Cefpirome | 30 | 5 | - | - |
| 10 | Cefpodoxime | 10 | 5 | - | - |
| 11 | Ceftazidime | 30 | 5 | - | - |
| 12 | Ceftazidime/Tazo | 30/10 | - | 1 | 4 |
| 13 | Ceftizoxime | 30 | 3 | 2 | - |
| 14 | Cefuroxime | 30 | 5 | - | - |
| 15 | Cephalothin | 30 | 1 | 4 | - |
| 16 | Cefotaxime | 10 | 5 | - | - |
| 17 | Ciprofloxacin | 5 | - | - | 5 |
| 18 | Co Trimoxazole | 25 | - | 1 | 4 |
| 19 | Doxycycline | 30 | - | - | 5 |
| 20 | Erythromycin | 15 | - | - | 5 |
| 21 | Ertapenem | 10 | - | - | 5 |
| 22 | Gentamycin | 10 | - | 2 | 3 |
| 23 | Imipenem | 10 | - | - | 5 |
| 24 | Levofloxacin | 5 | - | - | 5 |
| 25 | Linezolid | 30 | - | - | 5 |
| 26 | Methicillin | 5 | 5 | - | - |
| 27 | Meropenem | 10 | - | 3 | 2 |
| 28 | Moxifloxacin | 5 | - | - | 5 |
| 29 | Ofloxacin | 5 | - | - | 5 |
| 30 | Penicillin | 10 | 5 | - | - |
| 31 | Piper/Tazobactam | 100/10 | - | - | 5 |
| 32 | Teicoplanin | 30 | - | 2 | 3 |
| 33 | Tetracycline | 30 | - | - | 5 |
| 34 | Trimethoprim | 25 | 5 | - | - |
| 35 | Vancomycin | 30 | 3 | - | 2 |
Disc Diffusion Pattern of Enterococcus faeciuma
5. Discussion
Staphylococcus aureus and Enterococcus faecium have emerged as important pathogens, requiring bacterial anti-microbial therapy. These two strain’s resistance to various antibiotics is becoming increasingly frequent and resulting in serious therapeutic difficulties (6). Staphylococcus aureus and Enterococcus faecium appear to become drug resistant more readily than most other bacteria. In this study, Staphylococcus aureus and Enterococcus faecium strains were resistant to ampicillin, ampicillin sulbactam, methicillin, penicillin, cefixime, ceftazidime and trimethoprim may be due to no inactivation of the antibiotic as a result of structural modification by enzymatic action. The staphylococci and enterococci appear to become drug resistant more readily than most other bacteria. The appearance of drug resistant strains isolated from pathological processes has followed the introduction of various antibiotics in to general use and the proportion of resistant strains has continuously increased. Penicillin was the first antibiotic used for staphylococcal and enterococci infections, penicillin resistance appeared shortly after its introduction. This was followed by resistance to cotrimoxazole, ampicillin, amoxicillin and cefuroxime (7-9). In 1992, the first report of Staphylococcus aureus and Enterococcus faecium resistance to penicillin, aminopenicillin and antipseudomonal penicillin was published (10, 11). In general methicillin resistant staphylococcus aureusare multidrug resistant (12). In this study, three strains of Enterococcus faecium were resistant to vancomycin. More recent results from surveillance studies indicate that the proportion of Enterococcus faecium resistance to vancomycin continues to rise in patients hospitalized in the United States, approaching 70% in the most recent reports (13). In this study, linezolid and vancomycin antibiotics were sensitive to all Staphylococcus aureus strains. All the methicillin resistant Staphylococcus aureus were sensitive to linezolid, teicoplanin and vancomycin and Enterococcus faecium strains were also sensitive to linezolid and teicoplanin antibiotic (14). We report on multi-drug resistant staphylococci and enterococci and present an evaluation of laboratory tests for their detection. Both strains are still the most frequently detected pathogens, which have high resistance rates amongst bacteria isolated from hospital acquired infections especially in clinical impact of urine sample. Antibiotic resistance in Staphylococcus aureus and Enterococcus faeciumis becoming a persistent problem. Both infection control and antibiotic selective pressure are important factors contributing to the spread of these infections. The success and investment in antibiotic drug development would also be lost if appropriate measures are not put in place to stop the emergence of resistant strains of bacteria. The situation is more worrisome in less developed countries where inappropriate use of antibiotics is frequent. It is important to note that multi-drug resistant bacterial isolates may have sacrificed their pathogenic potential in order to acquire resistance to these antibiotics.