1. Background
Nowadays, hospital infections are a major problem around the world. These infections are important because of the increased length of hospitalization, diagnostic, treatment, and mortality rates. Various bacteria cause hospital infections. One of the most important bacteria that causes this infection is Escherichia coli. Bacteria are part of the normal flora of the intestine of humans and animals and most of them are opportunists pathogens (1).
Urinary tract infection (UTI) is one of the most common bacterial infections (2). Urinary tract infection is an important factor in the progressive destruction of kidney structure, chronic kidney failure, malformation, urolithiasis, and hypertension (3). Microbial infection of the urinary tract is one of the most common infectious diseases worldwide. The prevalence of this infection varies according to age and gender, and is clearly more pronounced in females than males due to anatomical differences, so that almost half of the females experience at least one urinary tract infection during their lifetime. Studies in different societies have suggested that gram-negative bacilli have been the most common etiologic factors in urinary tract infection and among them, E. coli accounts for more than 80% of cases of acute urinary tract infections (4).
Since the recognition of bacteria, humans have always been seeking effective drugs against infections caused by them and bacteria have developed effective mechanisms to eliminate antibiotics (5). Nowadays, with the advent of drug resistance among bacteria, the treatment of these infectious diseases has faced many problems (1).
Beta-lactams are considered as the most commonly used antibiotic group due to their low toxicity for eukaryotic cells, and extensive and strong antimicrobial effect. Beta-lactams are enzymes that hydrolyze the central nucleation of the beta-lactam ring, causing these antibiotics to be deactivated. One of the most important mechanisms of resistance to these antibiotics is the production of enzymes called extended spectrum beta-lactamases (ESBLs) and these factors are mainly located on the plasmid and could facilitate the expansion of these factors (6).
In the 1980s, Ambler classified these enzymes to four groups (A - D) based on the amino acid sequence, with type B having an active site on Zn2 + and a metallo-beta-lactamase.
OXa-type beta-lactamases are placed in the molecular D class and are able to disable oxazolyl of penicillin, such as dicloxacillin, cloxacillin, and oxacillin (7). In summary, several studies and reports indicate this type of resistance spread across the globe and this issue has become a global problem.
2. Objectives
The purpose of this study was to investigate the antimicrobial susceptibility of antibiotics used in the treatment of urinary tract infections caused by E. coli through the microdilution method and comparing it with disk diffusion.
3. Methods
The urine culture of the patients with urinary tract infections hospitalized at Imam Khomeini and Ali ibn Abi Talib hospital (Zabol, south-eastern Iran) were collected from September 2015 to March 2016. First, the samples were examined by microscopic Gram stain examination. Samples with gram-negative test results were inoculated on plates of nutrient agar, Clede agar, MacConkey's, and blood agar (Merck, Germany) and then incubated at 37°C for 24 hours. The test was performed as recommended by the clinical and laboratory standards institute (CLSI). Disks of amikacin (AN), cefazolin (CZ), amoxicillin-clavulanic acid (AMC), ampicillin (AM) were placed on Muller Hinton agar (MHA) at a distance of 30 mm between apart.
3.1. Minimum Inhibitory Concentration (MIC)
One of the earliest antimicrobial susceptibility testing methods was the microbroth or tube-dilution method (8). This procedure involved preparing two-fold dilutions of antibiotics (e.g. 1, 2, 4, 8, 16 , 32, 64, 128, 625, and 512 µg/mL) in a liquid growth medium dispensed in test tubes (8). The antibiotic-containing tubes were inoculated with a standardized bacterial suspension of 1 to 5 × 105 CFU/mL. Following overnight incubation at 35°C, the tubes were examined for visible bacterial growth as evidenced by turbidity. The lowest concentration of antibiotic that prevented growth represented the MIC. The precision of this method was considered to be plus or minus one 2-fold concentration, due, in large part, to the practice of manually preparing serial dilutions of the antibiotics (8). The advantage of this technique was the generation of a quantitative result (i.e. the MIC). The principal disadvantages of the macrodilution method were the tedious manual task of preparing the antibiotic solutions for each test, the possibility of errors in preparation of the antibiotic solutions, and the relatively large amount of reagents, and space required for each test.
3.2. Polymerase Chain Reaction
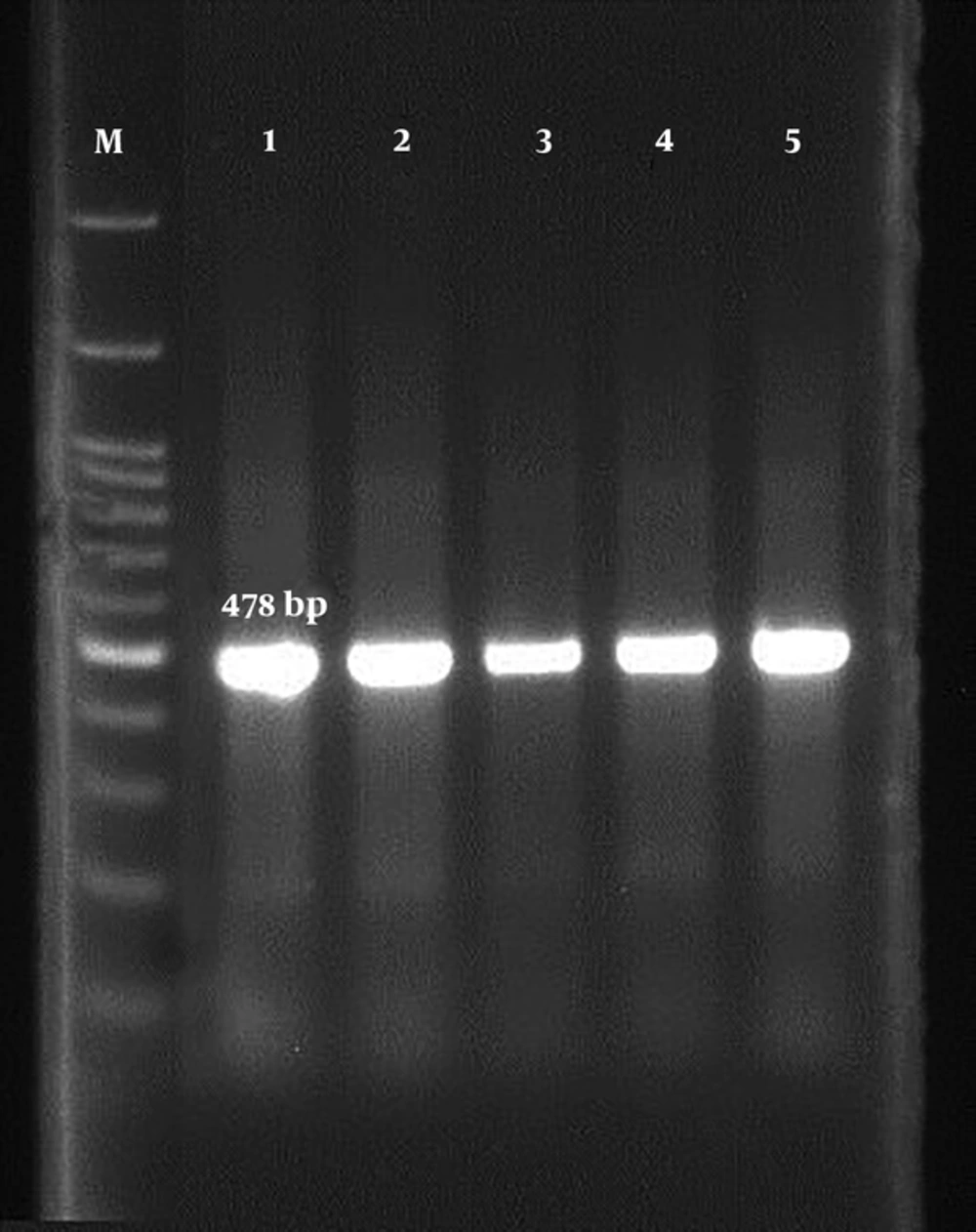
In order to detect OXA gene β-lactamase, genomic DNA of isolates was performed using the phenol-chloroform method. To identify the OxA beta-lactamase gene, OXA II F 5'-AAGAA ACGCTACTCGCCTGC-3'/ OXA II R 5'-CCACTCAACCCATC CTACCC-3 primer (478 bp) was used. Finally, a 25-μL PCR was performed, including 12 μL of Master mix, 1 μL of each primer, 3 μL of DNA and 8 μL of sterilized deionized water, for 35 cycles in the form of 15-minute cycles at 95°C. Then, three cycles consisting of a 30-second spin phase at 94°C, a connecting phase of 25 seconds at 59°C, a 60-second extension period at 72°C, and a 10-minute cycle at 72°C were carried out. The PCR product was electrophoresed in the presence of the gene in the 2% agarose gel and finally, they were stained with ethidium bromide and photographed and captured with ultraviolet (UV) light.
4. Results
The results of this study showed that E. coli was resistant to antibiotics, such as AN (42.85%), CZ (35.71%), AMC (35.71%), and AM (35.71%) (Table 1).
| AN | GM | AM | AMC | CZ | |
|---|---|---|---|---|---|
| R | 42.85 | 28.57 | 35.71 | 35.71 | 35.71 |
| S | 57.14 | 50 | 64.28 | 57.14 | 42.85 |
| I | 0 | 21.42 | 0 | 7.14 | 21.42 |
The results of this study showed that the MIC was for the antibiotic, cefazolin, which was equal to 16 μg/mL and two strains were inhibited at this concentration and the highest inhibitory concentration was 256 μg/mL. The lowest inhibitory concentration for ceftriaxone antibiotic was 8 μg/mL and the highest inhibitory concentration was 256 μg/mL. Five strains of E. coli grew at any concentration of cefotaxime antibiotics, while the highest inhibitory concentration was 128 μg/mL (Table 2). The results of PCR to detect the OXa gene showed that five strains (35.71%) were OXa gene carriers (Figure 1).
| Bacterial Code | Cefazolin | Ceftriaxone | Cefotaxime |
|---|---|---|---|
| 1 | 32 | 64 | Did not grow. |
| 2 | 64 | 64 | Did not grow. |
| 3 | 64 | 256 | Did not grow. |
| 4 | 64 | 64 | Did not grow. |
| 5 | 256 | 128 | Did not grow. |
| 6 | 64 | 128 | 128 |
| 7 | 64 | 128 | 128 |
| 8 | 64 | 32 | 64 |
| 9 | 128 | 32 | 64 |
| 10 | 64 | 32 | 32 |
| 11 | 128 | 32 | 32 |
| 12 | Grow | 16 | 32 |
| 13 | 16 | 8 | 16 |
| 14 | 16 | 32 | 8 |
5. Discussion
In the study of Ghadiri et al. (9), the pattern of antibiotic resistance of E. coli was studied in children. The MIC (50 and 90 percentiles) for each antibiotic was as fallow: ampicillin (256, 256), amikacin (1/5, 4/8), gentamycin (0.38, 32), nalidixic acid (1/5, 256), ceftriaxone (0.023, 32), cefixime (0.19, 256), and trimethoprim-sulfamethoxazole (32, 32). The antibiotic susceptibility rate for each antibiotic was as fallows: ampicillin (21.8%), amikacin (92%), gentamycin (75.5%), nalidixic acid (64.4%), ceftriaxone (72.4%), cefixime (65.5%), and trimethoprim-sulfamethoxazole (41.4%).
In contrast, the highest resistance rates were observed for ampicillin (77%), carbenicillin (76%), piperacillin (74%), and cotrimoxazole (62.5%), respectively (10).
Asadpour Rahimabadi et al. (11) tested the antibiotic resistance pattern in E. coli isolates in Rasht city. Their results showed that from 195 isolates of E. coli, 76.93% of isolates were from females and the rest were isolated from males. The highest sensitivity was for imipenem. The highest level of resistance in the penicillin family was related to oxacillin and ampicillin and in cephalosporins was related to cephalothin and among the quinolones, the highest resistance was related to nalidixic acid. On the other hand, the lowest resistance to gentamicin-nitrofurantoin and cefocytosine was 8.2% to 8.71%, and 11.79%, respectively.
Aghamahdi et al. at the 17th Shahrivar hospital of Rasht reviewed the incidence of UTI in the age of under two years old during years 2011 to 2006 (12). Urinary tract infection was higher in uncircumcised males, yet after six months it was more common in females. Therefore, male gender is a risk factor for UTI during infancy.
Abdollahi Kheirabadi et al. (13) examined the pattern of drug resistance in E. coli at Vali-e-Asr hospital of Fasa. The results indicated that the resistance of E. coli isolates to ciprofloxacin and imipenem was 22.7% and 11.1%, respectively according to the antibiogram test. The resistance of these strains was tested in a minimum concentration of inhibitor by dilution of 19.7% for ciprofloxacin and 9.4% for imipenem, relative to the entire sample.
Mollahosseini et al. (14) examined the pattern of antibiotic resistance of E. coli in patients of Shahid Sadoughi hospital in Yazd. The results showed that 104 samples (1.67%) of E. coli were isolated from microbiological and chemical tests. After using nine antibiotics, 1.34% gentamicin and 2.69% maroponin had the least resistance and the highest resistance was 20.62% and cefixime was 22.42%.
In another study, the E. coli were highly resistant to penicillin (100%), amoxicillin (100%) and cefotaxime (89.7%), followed by intermediate level of resistance to ceftazidime (73.8%), cephradine (73.8%), tetracycline (69.4%), doxycycline (66.6%), augmentin (62.6%), gentamycin (59.8%), cefuroxime (58.2%), ciprofloxacin (54.2%), cefaclor (50%), aztreonam (44.8%), ceftriaxone (43.3%), and imipenem (43.3%) (15).
The result of the study of Ferdosi-Shahandashti et al. (16) showed that E. coli grew in 57 urine samples. Imipenem, ofloxacin, and ciprofloxacin were the most sensitive antibiotics at 87.7%, 87.7% and 78.9% respectively. However, cotrimoxazole, cefixime, cefotaxime, and ceftriaxone were the most resistant antibiotics. Antibiotic sensitivity of disk diffusion compared to MIC detected by microdilution had sensitivity, specificity, positive predictive value, and negative predictive value of 82%, 98%, 99%, and 74%, respectively.
The result of the study of Akond et al. (17) show that 88%, 82%, 80%, 76%, 70%, 68%, 64%, 58%, 52%, and 20% of the tested E. coli strains from poultry sources were found resistant to penicillin, ciprofloxacin, rifampicin, kanamycin, streptomycin, cefixime, erythromycin, ampicillin, tetracycline, chloramphenicol and neomycin, respectively. None of the strains showed resistance to norfloxacin and gentamicin. Sensitivity was recorded in 86%, 80%, 60%, 36%, 30%, and 26% of strains to norfloxacin, gentamicin and chloramphenicol, neomycin, tetracycline, streptomycin and ampicillin, respectively.
In the study of 746 isolates recovered from animal sources, 531 (71.1%) were resistant to tetracycline, 441 (59%) to streptomycin, 431 (57.7%) to sulfonamide, 277 (37.1%) to kanamycin, and 255 (34.1%) to ampicillin. Among animal E. coli isolates, the rate of resistance was significantly higher in cattle isolates than in pig isolates for chloramphenicol (P = 0.039), amoxicillin/clavulanic acid (P = 0.03), sulfonamide (P = 0.038), and trimethoprim/sulfamethoxazole (P = 0.022). There was a significant difference in resistance rate between cattle and chicken isolates to ceftiofur (P = 0.008), ceftriaxone (P = 0.008), chloramphenicol (P = 0.011), and kanamycin (P = 0.037) (18).
The study of Chaudhary and Payasi (19) showed a great diversity of occurrence of oxacillinase (OXA) genes among clinical isolates. OXA-48 and OXA-10 were more prevalent in both E. coli (32.6% OXA-48; 16.3% OXA-10) and P. aeruginosa (OXA-48 32.4%; 27.0%) as evident by PCR. The incidence of other OXA genes in E. coli and P. aeruginosa varied from 4.0 to 12.1%. of tested drugs; cefepime plus sulbactam was found to be the most efficacious antibacterial agent with 81.6% to 89.2% susceptibility. Cefepime plus tazobactam was the second most active antibacterial agent with 46.9% to 56.7% susceptibility.
Knowledge of the spread of OXA-carbapenems in enteric bacteria is important as none of the currently available phenotypic tests (except super carba medium) is suitable for detecting OXA-48/OXA-181 producers (20). The activity of these enzymes is not inhibited by clavulanic acid, tazobactam, sulbactam, boronic acid, EDTA or dipicolinic acid (21).
The OXA-carbapenems activity with an increasing rate of ESBL activities of enteric bacteria in the hospital environment results in the development of multidrug resistance activity (22, 23). Carbapenems are supposed to be the last option for treatment, and the effectiveness of carbapenems has diminished (22, 24).