1. Background
Salmonella is one of the most important genera in Enterobacteriaceae family, which was first identified by Daniel Salmon (1). Many of the bacteria that constitute this genus can be pathogenic to humans and animals (2). These bacteria are among the most commonly transmitted bacteria from animals to humans. Due to their numerous animal reservoirs, this genus is one of the most important causes of foodborne diseases and is a health problem worldwide (3).
There is usually no cure for Salmonella infection in cases of acute and invasive disease, typhoid fever and immunodeficiency (4). According to the Center for Disease Control (CDC), currently 50% of Salmonella species are resistant to multiple drugs. Despite reports on resistance to various fluoroquinolones and cephalosporins in most parts of the world, these two drugs are nowadays the most effective drugs in the treatment of stem cells (5).
Recently, resistance to extended-spectrum cephalosporins such as cefotaxime, ceftriaxone, ceftazidime and ceftizoxime, which are mainly produced by β-lactamase, is increasing. Probably the excessive use of new antibiotics for treatment and selective pressure on bacteria has caused the production of new beta-lactamases by bacteria (6).
The extended spectrum of cephalosporin resistance is mainly related to the production of an enzyme called extended-spectrum beta-lactamase (ESBL). The ESBL enzyme is capable of hydrolyzing and disabling a wide range of β-lactams, including third-generation cephalosporins, penicillin, and aztreonam. This enzyme is the result of mutations in TEM-1, TEM-2, and SHV-1. All these beta-lactams are found in the Enterobacteriaceae family (7).
2. Objectives
This enzyme mediates resistance to extended spectrum cephalosporins and monobactams, such as aztreonam, but there is no detectable activity against cefixime and imipenem. Due to the extended range of substrates, these were called ESBL enzymes. The purpose of this study was to isolate CTX-M1 and CTX-M3 genes producing ESBL in clinical samples of Salmonella typhimurium isolated from poultry in Zabol, Iran.
3. Methods
3.1. Isolation of Bacteria
Samples of poultry feces were collected from Zabol, southeastern Iran, from September 2010 to March 2011. In brief, 1 mL of the sample from the transport swab was inoculated in 9 mL of buffered peptone water (Hi Media) and incubated at 37°C for 18 hours for pre-enrichment. Further, for selective enrichment 0.1 mL of the pre-enriched inoculum was transferred to 10 mL of Rappaport-Vassiliadis broth (Hi Media) and incubated at 42°C for 24 hours. After enrichment, a loopful (10 µL) of inoculums was then streaked on xylose lysine desoxycholate (XLD) agar (Hi Media) and incubated at 37°C for 24 hours. The presumptive Salmonella colonies (4 - 5 colonies/plate) appearing slightly transparent red halo with a black center surrounded by a pink-red zone on XLD agar were screened further for its biochemical characterization. The presumptive colonies of Salmonella were further subjected to biochemical tests viz., triple sugar iron (TSI), ortho-nitrophenyl galactosidase (ONPG), urease broth, indole, methyl red, Voges-Proskauer and Citrate test (IMViC) as per the standard test protocol described in bacteriological analytical manual FDA.
3.2. Preparation of 0.5 McFarland Suspension
A 0.5 McFarland standard is prepared by mixing 0.05 mL of 1.175% barium chloride dihydrate (BaCl2•2H2O), with 9.95 mL of 1% sulfuric acid (H2SO4) (8).
In order to prepare a microbial suspension, 24 hours before the experiment, bacterial culture was inoculated onto a sloped agar culture medium. After the growth of bacterial colonies, the culture medium was washed with normal saline solution and a concentrated microbial suspension was obtained. Then, some of the bacterial suspension was poured into a sterile tube containing normal saline solution and its opacity was measured with a spectrophotometer (Unico, US) at a wavelength of 630 nm. Afterwards, it was diluted with normal saline solution until the opacity of the solution was equal to 0.5 McFarland. Thus, bacterial suspension was prepared at a concentration of 1 × 108 CFU/mL.
3.3. Determining the Sensitivity of Bacteria to Conventional Antibiotics
The susceptibility of 24 bacterial strains to ampicillin (AM), gentamycin , cefazolin (CZ), amoxi-clav (AMC), and azithromycin (AZM) antibiotics (Antibody Medicine, Iran) was evaluated using standard kirby-bauer diffusion method. To this end, all bacterial strains with a concentration of 0.5 McFarland were prepared in a Mueller-Hinton agar. Antibiotic discs were placed at a proper distance from each other. The plates were incubated for 24 hours at 37°C and the diameter of the inhibitory zones was measured to determine the resistance and sensitivity of the strains to the desired antibiotics.
3.4. PCR Reaction
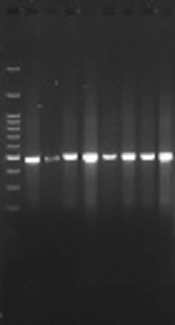
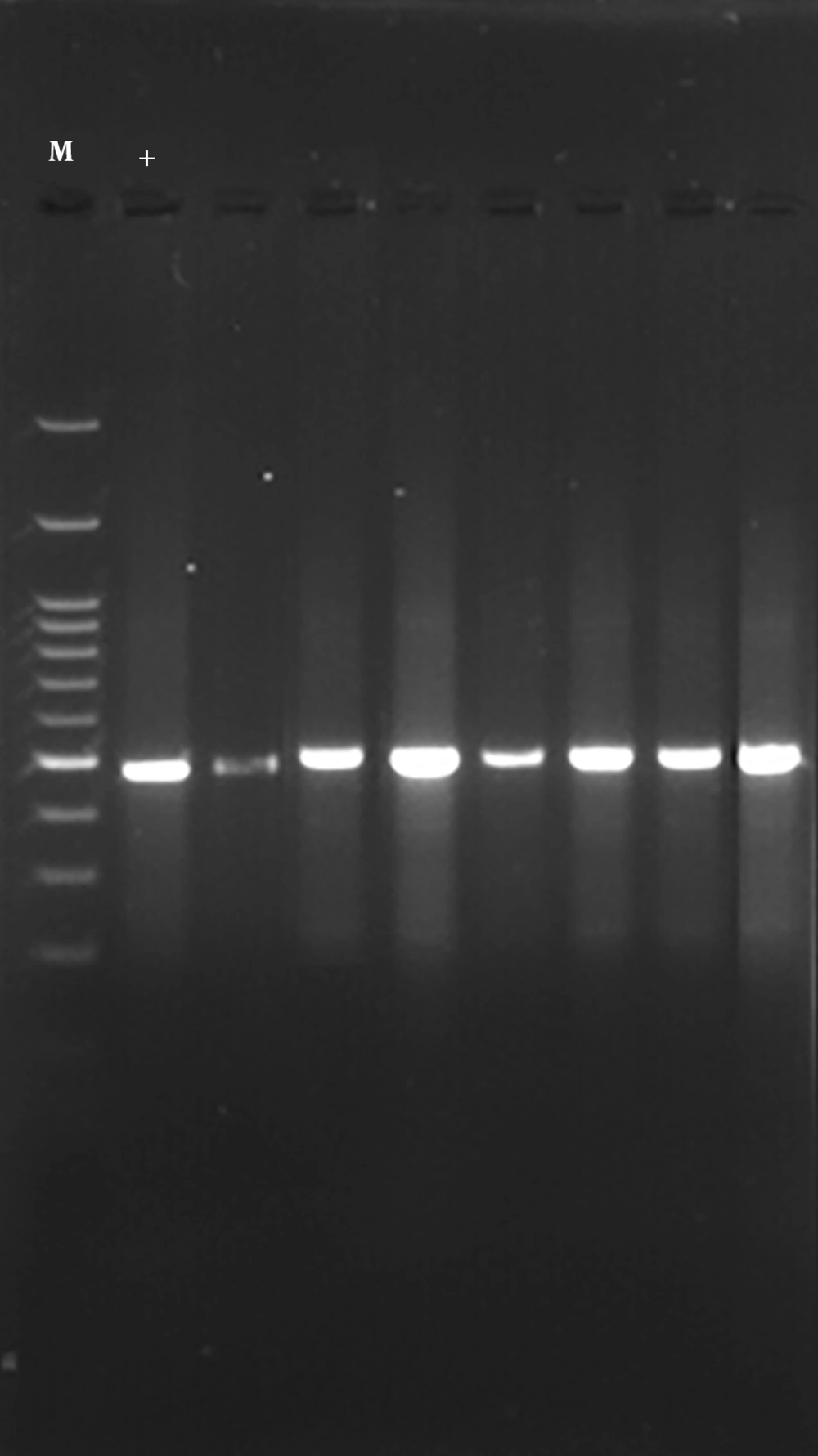
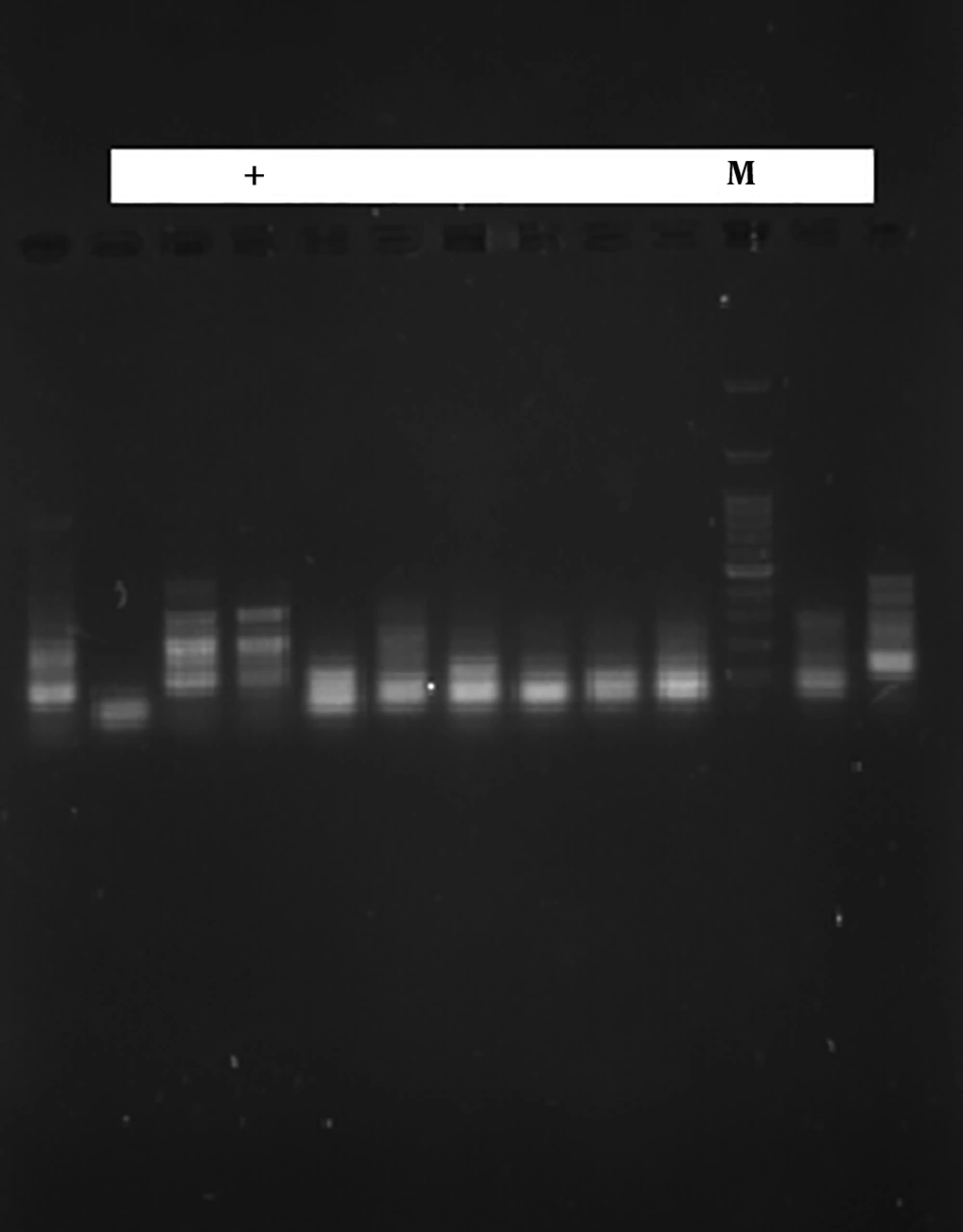
In order to detect the CTX-M1 and CTX-M3 genes of β-lactamase, genomic DNA of the isolates was identified using phenol-chloroform method. The identification of beta-lactamase genes, SHV, CTX-M1 and CTX-M3, was performed using the primers mentioned in Table 1 (9). Finally, the PCR reaction was carried out in a volume of 25 μL containing 12 μL Master Mix, 1 μL of each primer, 3 μL of DNA and 8 μL of sterilized deionized water, according to the primer temperatures presented in Table 2. The PCR product was electrophoresed in the presence of the gene on 2% agarose gel. Finally, the PCR product was stained with ethidium bromide and photographed and captured with ultraviolet (UV).
| Gene Name | Primer |
|---|---|
| CTX-M1 | |
| F | GACGATGTCACTGGCTGAGC |
| R | AGCCGCCGACGCTAATACA |
| CTX-M3 | |
| F | CGCTTTGCCATGTGCAGCACC |
| R | GCTCAGTACGATCGAGCC |
| Primer | PCR Reaction Steps | Number of Cycles | Reaction Product | ||
|---|---|---|---|---|---|
| Denaturation | Annealing | Extension | |||
| CTX-M1 | 35 | 499 | |||
| 94 | 94 | 55 | |||
| 180 s | 60 s | 30 s | |||
| CTX-M3 | 35 | 307 | |||
| 94 | 94 | 55 | |||
| 180 s | 60 s | 30 s | |||
4. Results
The results of this study showed that Salmonella typhimurium strains were resistant to AM (41.66%), GM (0%), CZ (0%), AMC (10%) and AZM (5%) antibiotics (Table 3).
| AM | GM | CZ | AMC | AZM | |
|---|---|---|---|---|---|
| S | 25 | 100 | 58.33 | 75 | 83.0 |
| I | 33.33 | 0 | 41.66 | 15 | 16.0 |
| R | 41.66 | 0 | 0 | 10 | 5 |
Abbreviations: AM, Ampicillin; AMC, amoxi-clav; AZM, azithromycin; CZ, cefazolin; GM, gentamycin.
The results of this study showed that of the 24 samples tested, 8 (28.57%) were positive for the CTX-M1 gene (Figure 1) and 12 (42.85%) were positive for CTX-M3 gene (Figure 2). These results indicate the low incidence of CTX-M1 and CTX-M3 genes in S. typhimurium.
5. Discussion
Salmonella species are considered as one of the most important food and gastroenteritis contaminants in humans. In Salmonella, there are more than 2600 serotypes, many of which are important pathogens for humans and animals (10, 11).
The World Health Organization has reported an annual Salmonella infection rate of 16 to 33 million patients and 500 to 600,000 deaths from Salmonella. It is a major health problem in the developing countries, including Iran (12).
On the other hand, the presence of carriers, which are usually difficult to detect and identify with the usual laboratory methods, in humans and animals plays a special role in the epidemiology of the disease. In Iran, the second cause of diarrhea in humans after Shigella is Salmonella (13).
Resistance to β-lactam antibiotics is mainly mediated by a large number of β-lactamases (14). Beta-lactamase enzymes have been identified in Salmonella, which are coded by different genes. Approximately, 10 genes such as TEM, SHV, PSE, and OXA have been identified as beta-lactamases (15).
The results of this study showed that S. typhimurium samples were resistant to AM (41.66%), GM (0%), CZ (0%), AMC (10%), and AZM (5%) antibiotics. The results of this study showed that of the 12 samples tested, 6 (42.85%) were positive for the CTX-M1 gene and 4 (28.57%) were positive for the CTX-M3 gene. These results indicate the low incidence of CTX-M1 and CTX-M3 genes in S. typhimurium.
In a study by Ziech et al. who tested S. typhimurium resistance patterns, the results showed that of 98 species, 84 were multidrug resistant. The highest resistance rates were observed to nalidixic acid (95%), tetracycline (91%), ampicillin (45%), streptomycin 19%, and gentamycin 15% (16).
In a study by Akinyemi et al., 35 (25.9%) Salmonella strains were isolated and identified, 74.3% of which were S. typhimurium and 22.9% were S. parathypha. A total of 24 strains produced beta-lactamases. Meanwhile, CTX-M1 gene was detected in about 45.8% of Salmonella specimens. They reported that 81.8% of S. typhimurium strains carry the CTX-M1 gene, while 18.2% of S. enteritysis strains carry the CTX-M1 gene (17).
Warren et al. observed that 33.3% of the E. coli isolated from poultry in Britain had ESBL genes (18). Jahantigh and Ordoni examined the prevalence of β-lactamase genes in E. coli isolated from poultry with Coli-septicaemia. The results showed that the prevalence of CTX gene in E. coli isolated from the chickens was 20%, which had an equal prevalence of 10% in the liver and kidneys. In TEM gene examination, the frequency of 24.28% was observed using PCT on E. coli isolated from the liver and kidneys (19).
Asadi et al. conducted a study on 56 E. coli isolates from poultry fecal samples in Urmia, Iran. They found that 26 (46.4%) isolates had CTX-M gene and 15 (26.7%) isolates had TEM gene (20).
In the study of Hasannejad et al., blaTEM, blaCTX-M and blaSHV were detected in E. coli isolated from poultry by multiplex-PCR and their antibiotic susceptibility profiles were examined. The antibiotic susceptibility test results showed that the lowest and highest resistance rates were related to imipenem (0%) and aztreonam (77%), respectively, and 41 (68.3%), 28 (46.6%) and 0 (0.0%) strains were positive for CTX-M, TEM and SHV, respectively. Also, 16 (26%) isolates carried both TEM and CTX-M genes (21).
5.1. Conclusions
The widespread use of beta-lactam antibiotics has increased the spread of ESBL enzymes in Iran and throughout the world and the use of these antibiotics is becoming more and more limited. Therefore, the complete identification of ESBL by experiments, the restriction of the use of beta-lactam antibiotics can maintain the efficacy of beta-lactam antibiotics as much as possible.