1. Background
Febrile neutropenia (FN) is defined as a single oral temperature of ≥ 101°F or a temperature of ≥ 100.4°F for at least an hour with an absolute neutrophilic count (ANC) of < 1500/µL. It is a common cause of complications and mortality in oncology patients, especially those with hematologic malignancies (1, 2). Despite the availability of granulocyte colony-stimulating factors (G-CSF), FN can necessitate intensive treatment, delay the chemotherapy effectiveness, and cause life-threatening infections (3).
Bloodstream infection (BSI) is a leading cause of mortality among FN patients (1-3). Over the past decades, the bacteriological profile of BSI in FN patients has undergone significant changes. Traditionally, Gram-negative bacteria were known as the primary causative agent of BSI in FN patients. Throughout the 1980s and 1990s, there was a decreasing incidence of Gram-negative bacteria accompanied by a relative increase in Gram-positive infection; a change attributed to factors such as better management of Gram-negative infections, prescription of prophylactic fluoroquinolones, and increased use of intravenous catheters (4, 5). The current situation of distribution of bacteria in pediatric FN in our community is controversial. Several studies have recently reported a re-emergence of Gram-negative organisms during the past few years (6-8). However, Gram-positive organisms have been reported as the most common pathogens in Iranian FN patients (9, 10).
The same as the distribution of pathogens, each hospital has its own antibiotic resistance profile, which could change over different time periods (11, 12). A study conducted at the Shaukat Khanum Memorial Cancer Hospital and Research Center from 2003 to 2006 revealed that Pseudomonas aeruginosa strains’ resistance to ciprofloxacin increased about 50% during this period (13). Colonization with antibiotic-resistant agents can be associated with the increased risk of infection in oncology patients (14). It has been shown that 5% - 50% of pediatric oncologic patients who have been colonized will develop bacteremia (15). Colonization with resistant pathogens in oncology patients may indicate a change in the FN management protocol, including antibiotics’ empiric administration (16).
Since the advent of empiric administration of broad-spectrum antibiotics at fever onset in neutropenic patients, guidelines have emphasized that the timely diagnosis and early initiation of supportive care and empiric broad-spectrum antibiotics are mandatory to prevent complications in FN patients (17-20). Choosing the best antibiotic regimen in FN increases empiric antibiotic therapy’s efficacy and decreases morbidities and health care costs. An updated distribution knowledge of prevalent pathogens specific to each center is required (9, 21).
2. Objectives
The periodic local investigation of bacterial profiles and antimicrobial sensitivity plays a crucial role in managing FN and reducing morbidity and mortality among pediatric oncology patients. This study aimed to investigate the distribution of bacterial pathogens and their sensitivity pattern in children with hematologic malignancies and FN. The secondary objective was to compare FN children’s colonization in the nasal, inguinal, and axillary regions on admission and 72 hours later to detect nosocomial colonization.
3. Methods
This prospective, cross-sectional study was carried out from December 2018 until the end of May 2019. The study population included children with hematologic malignancies admitted to the Amir Hematology and Oncology Hospital, a specialized and academic hospital affiliated to the Shiraz University of Medical Sciences, Shiraz, Iran.
The sample size was calculated according to the prevalence of positive blood cultures in FN patients, reported as 28.4% by Meidani et al. (9) and with the formula P (1-P) Z2/d2. A total of 78 FN children [defined as an oral temperature of ≥ 38.3°C with an absolute neutrophilic count (ANC) of < 500 cells/microL) aged ≤ 18 years were included. Of the participants, 13 children whose parents were unwilling to participate in the study were excluded, and the remaining 65 children were enrolled based on the inclusion criteria. The study participants were assured that participation in the project had no additional cost and refusal to participate in the project had no effect on the continuation of their treatment process. Informed consent forms were taken from the participants’ parents/guardians. The Ethics Committee approved the study protocol of the Medical Faculty at the Shiraz University of Medical Sciences, Shiraz, Iran (ethical code: 93-01-01-bb24).
Initially, the study outline was explained for the included patients and their families. Each patient’s nasal, axillary, and inguinal (NAI) areas were swabbed, and blood samples were taken. NAI cultures were repeated after 72 hours of admission to track nosocomial colonization. In case of fever continuation, deterioration of clinical conditions, or provision of a plan to change the antibiotic regimen, blood culture was repeated. A demographic questionnaire on the patients’ age, sex, and admission time, was filled out for each patient.
Blood cultures were carried out with BACTEC 9240 (Becton-Dickinson, US). Positive blood cultures prepared in the Cary-Blair transport medium (CONDA, Spain) were sent to Professor Alborzi at the” “Clinical Microbiology Research Center” for subcultures. Morphologic and biochemical characteristics identified all Gram-positive and -negative organisms. Catalase, Coagolase, and DNAase were used to identify Staphylococcus spp., and sensitivity to optochin was used to identify Streptococcus pneumoniae. Gram-negative organisms were identified based on colony morphology and biochemical characteristics using Microgen Kit (UK). Enterococcus was identified with growth on NaCl 6.5% and the Bile-Esculin Agar. To identify the extended-spectrum beta-lactamase (ESBL) producing organisms, double disks (CA2+clav, CTX-clav) were used. Staphylococcus aureus was isolated using the disk diffusion method with the cefoxitin disk. Antibiotic susceptibility tests were performed on Mueller Hinton Agar (Mast Diagnostics, Merseyside, UK) based on the 29th edition of the Clinical and Laboratory Standards Institute (CLSI) for performance standards of antimicrobial susceptibility testing (22). The presence of anaerobic bacteria was not investigated.
The data were analyzed using SPSS software, version 21.0. Descriptive statistics such as mean, standard deviation, and median were applied to describe the findings. To analyze the relationship between the studied variables, chi-square and Fisher’s exact test were used. P-value < 0.05 was considered as statistically significant.
4. Results
From December 2018 to June 2019, a total of 65 children (60% boys, 40% girls, mean age 7.3 ± 5.3 years) with hematological malignancies were diagnosed with FN and admitted to the Amir Hematology and Oncology Hospital. All the patients were aged 18 years or younger and had an ANC < 500 cells/microL. The most common cause of admission in the patients was fever. Acute lymphoblastic leukemia (ALL) was the most common underlying hematological malignancy reported in 37 of the patients (56.9%). Acute myeloid leukemia (AML), non-Hodgkin lymphoma (NHL), and Hodgkin lymphoma (HL) accounted for the rest of the patients. Fifty-five (84.6%) of the patients had a history of the previous admission. Of them, 50 patients had been hospitalized during the previous month. Totally, 40 of the patients (61.5%) had a history of neutropenia before the current admission.
According to the results of the cultures, the most common grown bacteria in the NAI samples were coagulase negative staphylococci (CoNS), ranging from 29 (44.6%) of the nasal samples to 52 (80%) of the axillary samples. In the second set of NAI samples obtained after 72 hours, similar culture results were observed, except for one case of Enterococcus spp. in the axillary samples and two Proteus spp cases in nasal and inguinal samples (Table 1).
| Culture Area | On Admission | 72 Hours After Admission |
|---|---|---|
| Inguinal | ||
| CoNS | 42 (64.6) | 40 (61.5) |
| Staphylococcus aureus | 3 (4.6) | 1 (1.5) |
| Enterococcus spp. | 5 (7.7) | 3 (4.6) |
| ESBL-producing E. coli | 3 (4.6) | 3 (4.6) |
| Non-ESBL-producing E. coli | 3 (4.6) | 1 (1.5) |
| ESBL-producing Klebsiella | 1 (1.5) | 3 (4.6) |
| Proteus spp. | 0 (0) | 4 (6.2) |
| Negative culture | 8 (12.4) | 10 (15.5) |
| Nose | ||
| CoNS | 29 (44.6) | 32 (49.3) |
| Staphylococcus aureus | 15 (23.2) | 16 (24.6) |
| Enterococcus spp. | 1 (1.5) | 0 (0) |
| Streptococcus pneumoniae | 1 (1.5) | 0 (0) |
| Proteus spp. | 0 (0) | 1(1.5) |
| Negative culture | 19 (29.2) | 16 (24.6) |
| Axillary | ||
| CoNS | 52 (80.0) | 50 (76.9) |
| Staphylococcus aureus | 4 (6.2) | 3 (4.6) |
| Enterococcus spp. | 0 (0) | 1 (1.5) |
| Negative culture | 9 (13.8) | 11 (17) |
Abbreviations: CoNS, coagulase-negative staphylococci; ESBL, extended-spectrum beta-lactamases.
aValues are expressed as No. (%).
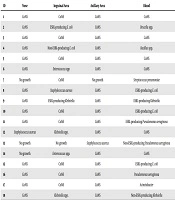
As shown in Table 2, 18 out of the 65 FN patients (27.7%) had positive blood cultures, with 11 (61.1%) of the isolated pathogens being Gram-negative and seven (38.9%) Gram-positive. Overall, the most common Gram-negative bacteria were E. coli and Pseudomonas aeruginosa, each constituting 27.2% of the Gram-negative isolates. CONS constituted six out of the Gram-positive isolates (85.7%).
Finally, we compared the results of the environmental cultures with those of the blood cultures. As shown in Table 2, the blood cultures’ results did not match those of the NAI cultures in 11 out of 18 patients with positive blood cultures (61.1%). Positive blood cultures were also examined for antimicrobial sensitivity and resistance (Table 3).
| ID | Nose | Inguinal Area | Axillary Area | Blood |
|---|---|---|---|---|
| 1 | CoNS | CoNS | CoNS | CoNS |
| 2 | CoNS | ESBL-producing E. coli | CoNS | Brucella spp. |
| 3 | CoNS | CoNS | CoNS | CoNS |
| 4 | CoNS | Non-ESBL-producing E. coli | CoNS | Bacillus spp. |
| 5 | CoNS | CoNS | CoNS | CoNS |
| 6 | CoNS | Enterococcus spp | CoNS | CoNS |
| 7 | No growth | CoNS | No growth | Streptococcus pneumoniae |
| 8 | CoNS | Staphylococcus aureus | CoNS | ESBL-producing E. coli |
| 9 | CoNS | ESBL-producing Klebsiella | CoNS | ESBL-producing Klebsiella |
| 10 | CoNS | CoNS | CoNS | ESBL-producing E. coli |
| 11 | CoNS | CoNS | CoNS | ESBL-producing Pseudomonas aeruginosa |
| 12 | Staphylococcus aureus | Klebsiella spp. | CoNS | CoNS |
| 13 | No growth | No growth | Staphylococcus aureus | Non-ESBL-producing Pseudomonas aeruginosa |
| 14 | No growth | Enterococcus spp. | CoNS | CoNS |
| 15 | CoNS | CoNS | CoNS | ESBL-producing E. coli |
| 16 | CoNS | CoNS | CoNS | Pseudomonas aeruginosa |
| 17 | CoNS | CoNS | CoNS | Acinetobacter |
| 18 | CoNS | Klebsiella spp. | CoNS | Non-ESBL-producing Klebsiella |
Abbreviations: CoNS, coagulase-negative staphylococci; ESBL, extended-spectrum beta-lactamases.
| Pathogen | Sensitive | Resistant |
|---|---|---|
| CoNS | Cefepime; Cefoxitin; Ciprofloxacin; Gentamicin; Imipenem; Linezolid; Rifampin; Vancomycin; Penicillin | Methicillin; Ceftazidime; Clindamycin; Colistin; Cotrimoxazole |
| ESBL-producing Klebseilla | Ceftazidime; Ciprofloxacin; Imipenem; Meropenem; Polymyxin B; Vancomycin; Amikacin | Ampicillin; Ampicillin-sulbactam; Ceftriaxone; Cefixime; Cephalexin; Colistin; Gentamicin; Cotrimoxazole |
| Non-ESBL-producing Klebsiella | Amikacin; Aztreonam; Ceftriaxone; Cefixime; Ciprofloxacin; Colistin; Gentamicin; Imipenem; Meropenem; olymyxin B | Ampicillin- sulbactam; Cotrimoxazole |
| Acinetobacter | Cefepime; Amikacin; Ciprofloxacin; Colistin; Gentamicin; Meropenem; Polymyxin B | Ampicillin; Aztreonam; Cephalexin; Cotrimoxazole |
| Non-ESBL-producing Pseudomonas aeruginosa | Ampicillin-sulbactam; Cefotaxime; Ceftriaxone; Ciprofloxacin; Colistin; Imipenem; Meropenem; Polymyxin B | Ampicillin; Amikacin; Ampicillin-sulbactam; Aztreonam; Cefixime; Cefuroxime; Cotrimoxazole; Cephalexin; Gentamicin |
| ESBL-producing E. coli | Amikacin; Aztreonam; Ceftazidime; Colistin; Imipenem; Meropenem; Polymyxin B | Cefepime; Ampicillin; Cefotaxime; Ceftriaxone; Cefuroxime; Cephalexin; Ciprofloxacin; Gentamicin; Cotrimoxazole |
| Streptococcus pneumoniae | Cefepime; Cefotaxime; Ceftriaxone; Ciprofloxacin; Clindamycin; Imipenem; Linezolid; Rifampin; Vancomycin | Cefoxitin; Cephalexin; Gentamicin |
| ESBL-producing Pseudomonas aeruginosa | Ciprofloxacin; Colistin; Imipenem; Polymyxin B | Cefepime; Ampicillin; Amikacin; Ampicillin-sulbactam; Aztreonam; Cefotaxime; Ceftriaxone; Ceftazidime; Cotrimoxazole; Gentamicin |
5. Discussion
Despite advances in FN management, infections remain a significant challenge in pediatric oncology (23). In the present study, blood culture was positive in about 28% (18 out of 65) of children with FN and underlying hematologic malignancies. Various BSI rates were reported by Zermatten et al. (21%), Mvalo et al. (26.9%), and Siddaiahgari et al. (40%), which may be explained by different diagnostic methods and procedures applied for the detection of infective agents (24-26).
In our study, Gram-negative bacteria constituted 61% of the isolates, with E. coli and Pseudomonas aeruginosa being the most frequently isolated organisms. CONS comprised 85.7% of the Gram-positive isolates. Siddaiahgari et al. (26) reported Gram-negative isolates in 85.3% of positive blood or urine cultures, with E. coli and Pseudomonas aeruginosa being the most common pathogens. Karanwal et al. (21) reported Gram-negative bacteria (mostly E. coli) in 78% of blood, sputum, and stool samples. Conversely, the predominance of Gram-positive organisms was reported by authors such as Meidani et al. (56.4%) and Lehrnbecher et al. (80.3%) (9, 27). The higher prevalence of Gram-negative organisms in the studies by Karanwal et al. (21) and Siddaiahgari et al. (26) might respectively be due to stool and urine specimens in their samples. On the other hand, prophylactic trimethoprim-sulfamethoxazole and the use of indwelling catheters might respectively have led to a lower rate of Gram-negative and higher rate of Gram positive infections in studies by Lehrnbecher et al. (27) and Meidani et al. (9).
According to our results, Gram-negative isolates from the blood cultures were mostly resistant to ampicillin (8, 72.7%), cotrimoxazole (6, 54.5%), cefepime (5, 45.4%), cefixime (3, 27.3%), and ceftriaxone (2, 18.2%) whereas Gram-positive isolates were mostly resistant to clindamycin (4, 57.1%), cotrimoxazole (4, 57.1%), and methicillin (3, 42.9%). Half of the CONS were methicillin-resistant. This pattern may be due to the overprescription of these antibiotics in our country. In a study from Uganda, Gram-negative organisms were isolated from febrile oncology patients, and Enterobacteriaceae were resistant to cotrimoxazole, ceftriaxone, and ampicillin (28). We also found that ESBL-producing E. coli and ESBL-producing Pseudomonas aeruginosa, accounting for about fourth of the pathogens, were resistant to cefepime which is currently used as the primary empirical therapy for FN management at our healthcare center. Further studies are needed to clarify the pattern of bacterial resistances and warrant update for FN guideline.
Furthermore, we assessed the colonization pattern in various skin sites, including axillary, inguinal, and nasal regions on admission and 72 hours later in hospitalized FN children. The most common colonized organisms were CoNS in the inguinal and nasal areas and MR CoNS in the axillary area. However, after 72 hours, ESBL-producing Klebsiella, Proteus, and Enterococcus were also isolated, which can be attributed to nosocomial colonization. Similarly, CoNS were the most common colonized pathogens in an analysis of 121 nasal, oropharyngeal, and anal swabs taken from children with chemotherapy-related FN by Spinardi et al. (15) The second common pathogen was Enterococcus, which may be due to rectal samples.
In our setting, comparing the blood and NAI cultures revealed no significant concordance; in about 60% of patients with bacteremia, the pathogen responsible for BSI was different from the colonized microorganism. Conversely, a study conducted in Chicago found that skin cultures were positive in all patients with vancomycin-resistant Enterococci-induced bacteremia, showing a significant association between colonization and BSI (29). Another study performed by von Eiff et al. (30) reported that most patients with Staphylococcus aureus-induced bacteremia had the pathogen colonization in their nasal cavity, which is discordant to our findings.
Our study’s main limitation was its small sample size, mainly resulting from the recruitment of patients from a single center and the relatively short duration of the study. Multi-center studies with a longer duration and larger sample size are warranted. Moreover, we recommend studies concerning the evaluation of the cost-effectiveness of different antibiotics and escalation therapy’s role in decision-making.
5.1. Conclusions
In our study, Gram-negative bacteria were the predominant etiologic agents of FN, with ESBL-producing E. coli being most frequently isolated. Most Gram-negative pathogens were resistant to cefepime. The results show a high resistance pattern in Gram negative pathogens.
Nasal, axillary and inquinal (NAI] colonization on admission and 72 hours later showed the isolation of new Gram-negative pathogens. No significant relationship was detected between the blood and NAI cultures. We recommend surveillance of microorganism colonization and antibiotic resistance patterns to select appropriate antibiotics for FN children’s empiric treatment.