1. Background
Acute respiratory tract infections (ARTI) are the most common cause of severe disease and mortality in children worldwide, especially in the first year of life (1). Most of these infections are caused by at least one of the following respiratory pathogens, including bacteria like mycoplasma (MP) or chlamydia (CH), viruses, and fungi (2). The identification of respiratory pathogens (RPs) not only paves the way for accurate diagnosis of ARTI but also is associated with the improved effect of therapy, declined hospitalization, and reduced quantity of antibiotics used in the hospital (3).
However, a timely and accurate diagnosis of respiratory pathogens can be challenging. Rapid antigen tests are the common way to detect the influenza virus, but they suffer from deficiencies in sensitivity, with the possibility of false-positive or negative results (2). However, increasing attention is being paid to multiplex-PCR technology, which in addition to its simplicity and lower operation time, can detect multiple RPs in a single sample. In addition, this assay allows detecting co-infections that may cause severe ARTI. The prevalence of ARTI in children varies depending on the region and age (4). However, limited information are available regarding the ARTI in Henan province
2. Objectives
To analyze the pathogens in hospitalized pediatric patients with ARTI in Henan province, we investigated the epidemiology of RPs via multiplex-PCR based on the 3500Dx Genetic Analyzer platform.
3. Methods
3.1. Sample Collection
In the present study, we retrospectively analyzed data of 11306 ARTI patients admitted to Henan Children's Hospital from March 1, 2019, to February 29, 2020. The specimens were transferred from the inpatient department to the laboratory and stored at 4°C for one day. Those younger than 15 years old and patients who were suffering from symptoms of ARTI according to the diagnostic criteria of “Zhu Futang Practical of Pediatrics” (ISBN 978-7-117-19978-O/R·19979) were included in the study. Children with ARTI had at least one of the following symptoms: cough, fever, sore throat, difficulty breathing, nasal congestion, or runny nose. Exclusion criteria were: 1). few cells in the specimen or insufficient amount of nucleic acid; and 2). Incomplete clinical data.
For all children, specimens were collected within 48h of hospitalization. A total of 11306 specimens were collected in this study. Depending on the patient’s status, sputum (n = 523; 4.6%), bronchoalveolar lavage fluid (n = 212; 1.9%) or pharyngeal swab (n = 10571; 93.5%) was obtained by sputum aspirator, bronchoscopy, or pharyngeal swabs (Shenzhen Mairuikelin Technologies Ltd., Shenzhen, China), respectively. Specimens were transferred from the inpatient department to the laboratory and stored at 4 °C at most for one day.
3.2. Total Nucleic Acid Extraction
Nucleic acids were extracted using a throat swab extraction kit (Health Gene Technologies, China)and a Tanbead Nucleic Acid Extractor (Tanwan Advanced Nanotech Inc, China).
3.3. PCR
We used the Respiratory Pathogen Multiplex Detection Kit (Ningbo HEALTH Gene Technologies Ltd., Ningbo, China) for multiplex PCRs, which includes PCR enzyme, dNTPs, MgCl2, primers for each pathogen, and buffer. Multiplex-PCR mixtures contained 14 μL of the pre-mixed solution, 1 μL of enzyme mix (containing uracil-DNA glycosylase enzyme, reverse transcriptase, and DNA polymerase), and 5 μL of the extracted sample. Multiplex PCR was completed on a Verity thermal cycler (Thermo Fisher Scientific Inc) following this methodology: step 1, 25°C for 5 min, 50°C for 15 min, and 95°C for 2 min; step 2, 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. The second step was iterated for 35 cycles. Then, samples were incubated at 72°C for 10 min and afterward were stored at 4°C. The kit of the multiplex-PCR assay (Health Gene Technologies) can detect 12 pathogens, including human rhinovirus (HRV), human respiratory syncytial virus (HRSV), influenza A H3N2 (H3N2), human adenovirus (HADV), human parainfluenza virus (HPIV), human metapneumovirus (HMPV), influenza B (IFVB), human bocavirus (BOCA), human coronavirus (HCOV), influenza A H1N1(H1N1), CH, and MP.
3.4. Runing on the 3500Dx Genetic Analyzer Platform
Following the multiplex-PCR, 1μL of amplified products and 0.25μL of SIZE-500PLUS (Thermo Fisher Scientific Inc) were added into 8.75μL of Hi-DiTM Formamide (Thermo Fisher Scientific Inc). Then, analysis was performed on the mixed liquid using the 3500DX Genetic Analyzer platform (Thermo Fisher Scientific, USA).
3.5. Statistical Analysis
Raw data were analyzed using SPSS software (version 23.0; IBM Corp., USA). Positive rates among different age groups and seasons were analyzed using the chi-squared test. Statistical significance was considered when P-value < 0.05.
4. Results
4.1. Confirmation of the Assay Based on the 3500DX Genetic Analyzer Platform
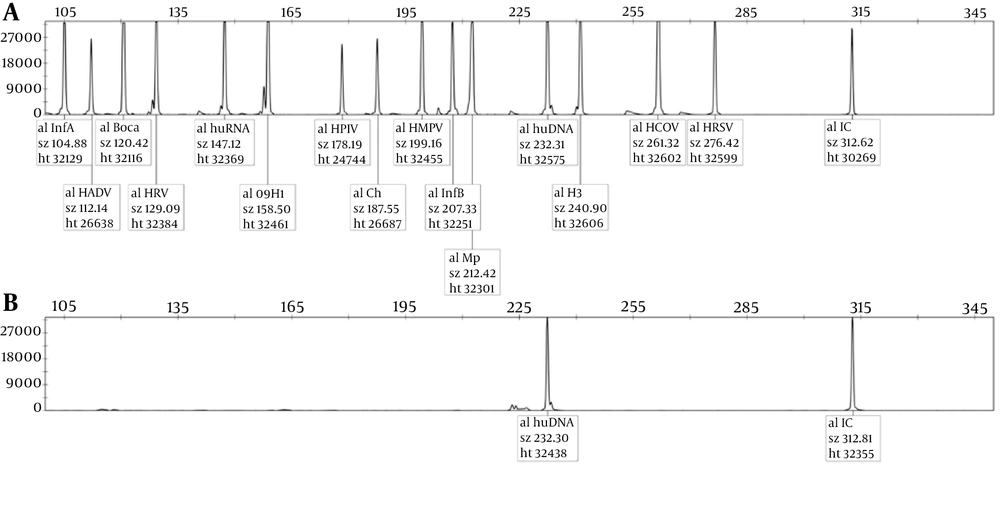
Human genomic DNA was used as a negative control, and plasmids carrying the corresponding viral sequences were used as a positive control. The human DNA peak and internal reference were observed in the negative control, while 12 pathogen-specific peaks presented in the positive control, and the peaks separated well. We found no overlap in the positive control (Figure 1), which indicates the accuracy and reliability of the assay based on the 3500DX Genetic Analyzer platform.
4.2. Characteristics of the Patients
A total of 11306 children with ARTI (6650 boys and 4656 girls) who were hospitalized from March 1, 2019, to February 29, 2020, were included in the present study. The youngest and oldest participants aged 4 days and 15 years, respectively. The general characteristics of participants are provided in Table 1.
| Characteristics | No. (%) |
|---|---|
| Gender | |
| Male | 6650 (58.8) |
| Female | 4656 (41.2) |
| Age (y) | |
| < 1 | 2944 (26.0) |
| 1 - 3 | 3371 (29.8) |
| 3 - 5 y | 2542 (22.5) |
| 5 - 15 y | 2449 (21.7) |
| Total | 11306 |
4.3. Results of Respiratory Pathogen Detection
The overall positive rate was 78.1% (8831/11306). Of 8831 positive samples, 7017 (79.5%, 7017/8831) had a single pathogen and 1814 (20.5%, 1814/8831) had multiple pathogens. The rate of single infection was 62.5% (7071/11306) and that of multiple infections was 16.0% (1814/11306). HRV was the most common pathogen (25.4%, 2874/11306), followed by MP (18.1%, 2050/11306), and HRSV (15.8%, 1783/11306). The positivity rates of other pathogens were as follows: IFVA-H3N2, 9.0% (1023/11306); HADV, 8.7% (982/11306); HPIV,7.3% (820/11306); HMPV, 3.3% (371/11306); IFVB, 3.2% (367/11306); BOCA, 2.0% (231/11306); HCOV,1.9% (220/11306); Ch,0.4% (46/11306); and IFVA-H1N1, 0.1% (13/11306).
4.4. Age and Gender Distribution of Positive Patients
The positive rates of RPs in boys and girls were 78.5% (5219/6650) and 77.6% (3612/4656), respectively. There was no significant difference between boys and girls concerning the variable of positive rate (χ2 = 0.286, P = 0.593). In this study, using the variable of age, participants were categorized as follows: babies (< 1 year), toddlers (1 - 3 years), preschool children (3 - 5 years), and school children (5 - 15 years). The distribution of RPs in the four groups is shown in Table 2. There was no significant difference among the different groups concerning the total positive rates.
| Characteristics | Babies (< 1y) N = 2944 | Toddlers (1 - 3y) N = 3371 | Preschool Children (3 - 5y) N = 2542 | School Children(5 - 15y) N = 2449 | All Ages N = 11306 | Χ2 | P-Value | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No.Pos. | Prevalence (%) | No.Pos. | Prevalence (%) | No.Pos. | Prevalence (%) | No.Pos | Prevalence (%) | No.Pos | Prevalence (%) | |||
| Positive | 2211 | 75.1 | 2653 | 78.7 | 2071 | 81.5 | 1896 | 77.4 | 8831 | 78.1 | 7.4 | > 0.05 |
| Single | 1763 | 59.9 | 2032 | 60.3 | 1614 | 63.5 | 1610 | 65.7 | 7019 | 62.1 | 10.2 | < 0.05 |
| Mixed | 448 | 15.2 | 621 | 18.4 | 457 | 18.0 | 286 | 11.7 | 1812 | 16.0 | 48.2 | < 0.05 |
| HADV | 132 | 4.5 | 338 | 10.0 | 320 | 12.6 | 192 | 7.8 | 982 | 8.7 | 113.4 | < 0.05 |
| BOCA | 88 | 3.0 | 112 | 3.3 | 26 | 1.0 | 5 | 0.2 | 231 | 2.0 | 93.4 | < 0.05 |
| HRV | 811 | 27.5 | 946 | 28.1 | 691 | 27.2 | 426 | 17.4 | 2874 | 25.4 | 79.7 | < 0.05 |
| IFVB | 49 | 1.7 | 111 | 3.3 | 95 | 3.7 | 113 | 4.6 | 367 | 3.2 | 38.6 | < 0.05 |
| CH | 19 | 0.6 | 4 | 0.1 | 6 | 0.2 | 17 | 0.7 | 46 | 0.4 | 17.8 | < 0.05 |
| H1N1 | 2 | 0.1 | 6 | 0.2 | 3 | 0.1 | 2 | 0.1 | 13 | 0.1 | 2.0 | > 0.05 |
| MP | 114 | 3.9 | 396 | 11.7 | 534 | 21.0 | 1006 | 41.1 | 2050 | 18.1 | 1128.7 | < 0.05 |
| H3N2 | 155 | 5.3 | 274 | 8.1 | 309 | 12.2 | 285 | 11.6 | 1023 | 9.0 | 95.0 | < 0.05 |
| HCOV | 71 | 2.4 | 83 | 2.5 | 35 | 1.4 | 31 | 1.3 | 220 | 1.9 | 18.0 | < 0.05 |
| HRSV | 855 | 29.0 | 618 | 18.3 | 282 | 11.1 | 28 | 1.1 | 1783 | 15.8 | 710.4 | < 0.05 |
| HPIV | 310 | 10.5 | 319 | 9.5 | 140 | 5.5 | 51 | 2.1 | 820 | 7.3 | 167.2 | < 0.05 |
| HMPV | 95 | 3.2 | 128 | 3.8 | 110 | 4.3 | 38 | 1.6 | 371 | 3.3 | 33.6 | < 0.05 |
4.5. Seasonal Distribution of RPs
Participants were separated into four groups according to the season: (a) spring (February, March, and April) with 696 cases; (b) summer (May, June, and July) with 888 cases; (c) autumn (August, September, and October) with 3688 cases; and (d) winter (November, December, and January) with 6034 cases. The positive rates of RPs in every season are shown in Table 3. It is worth noting that HRSV was found to be more frequent during the winter, while the positive rate of HRV showed little change throughout the year. Also, MP was more common in autumn. The rest of the RPs had an almost equal incidence throughout the year, i.e., without a significant seasonal trend.
| Spring (696) | Summer (888) | Autumn (3688) | Winter (6034) | Total (11306) | Χ2 | P-Value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No.Pos. | Prevalence (%) | No.Pos. | Prevalence (%) | No.Pos. | Prevalence (%) | No.Pos. | Prevalence (%) | No.Pos. | Prevalence (%) | |||
| HADV | 38 | 5.5 | 99 | 11.1 | 401 | 10.9 | 444 | 7.4 | 982 | 8.7 | 54.0 | < 0.05 |
| BOCA | 4 | 0.6 | 42 | 4.7 | 105 | 2.8 | 80 | 1.3 | 231 | 2.0 | 63.0 | < 0.05 |
| HRV | 54 | 7.8 | 354 | 39.9 | 1453 | 39.4 | 1013 | 16.8 | 2874 | 25.4 | 561.3 | < 0.05 |
| IFVB | 55 | 7.9 | 29 | 3.3 | 10 | 0.3 | 273 | 4.5 | 367 | 3.2 | 220.9 | < 0.05 |
| CH | 3 | 0.4 | 4 | 0.5 | 19 | 0.5 | 20 | 0.3 | 46 | 0.4 | 4.0 | > 0.05 |
| H1N1 | 3 | 0.4 | 0 | 0.0 | 1 | 0.0 | 9 | 0.1 | 13 | 0.1 | 12.0 | < 0.05 |
| MP | 152 | 21.8 | 110 | 12.4 | 865 | 23.5 | 923 | 15.3 | 2050 | 18.1 | 212.5 | < 0.05 |
| H3N2 | 29 | 4.2 | 12 | 1.4 | 11 | 0.3 | 971 | 16.1 | 1023 | 9.0 | 715.0 | < 0.05 |
| HCOV | 2 | 0.3 | 40 | 4.5 | 96 | 2.6 | 82 | 1.4 | 220 | 1.9 | 52.9 | < 0.05 |
| HRSV | 72 | 10.3 | 0 | 0.0 | 59 | 1.6 | 1652 | 27.4 | 1783 | 15.8 | 1157.5 | < 0.05 |
| HPIV | 28 | 4.0 | 158 | 17.8 | 491 | 13.3 | 143 | 2.4 | 820 | 7.3 | 551.6 | < 0.05 |
| HMPV | 33 | 4.7 | 1 | 0.1 | 13 | 0.4 | 324 | 5.4 | 371 | 3.3 | 232.5 | < 0.05 |
5. Discussion
RPs are the direct cause of ARTI. Hence, their early detection not only can help clinicians to make an accurate diagnosis but also promotes rational prescription of antibiotics. In recent years, there has been an increase in the use of nucleic acid amplification tests (NAAT) for the detection of RPs due to their excellent detection ability (5).
In this study, we found significant differences concerning the distribution of RPs based on the season and age. It is well-proved that the most common pathogen depends on age and season. For instance, a study conducted in Iran reported that the most common respiratory pathogen was HRV in the group younger than 4 years and IFVB in the group of 5 to 50 years (6). Or a study in the United States mentioned BOCA as the most prevalent respiratory pathogen among those younger than 4 years and HRV in the group of 5 to 50 years (7). These reports suggest that the prevalence of RPs depends on the geographic regions, in addition to season and age.
Several studies analyzed the epidemiological patterns of respiratory infections. HRV is usually the most common single pathogen found in ARTI samples, with a prevalence of 24 - 50%, followed by HRSV (22 - 25%), and influenza viruses (7.2 - 8%), either in nasal washes or nasopharyngeal swabs (8-10). In this study, the most common RP was HRV (25.4%, 2874/11306), followed by MP (18.1%, 2050/11306) and HRSV (15.8%, 1783/11306).
According to the findings, gender had no effect on the prevalence positive rate of RPs, which is consistent with the literature (11-13). Many studies concluded that respiratory infections are influenced by the season, with their peaks lasting from winter to early spring (12, 14). In this study, the positive rate of RPs was higher in winter, while it was lower in the spring. Hence, it can be argued that climate has an important role in the prevalence of RPs, since cold air may extend the survival time of RPs in respiratory droplets. Huang et al. reported no significant difference concerning the total RPs positivity rate of different age groups. However, they mentioned that the spectrum of RPs was different depending on the age group (12), which is in line with the findings of the present study. This difference can be attributed to several factors like immunity, guardian status, and range of activity.
The global incidence of influenza ranges from 5 to 10% among adults and 20 to 30% among children (15). Influenza virus can cause severe pneumonia, which may even be fatal. According to the literature, influenza is disproportionate in its effect on different age groups, so that it mainly affects younger children (nearly 870 000 cases of hospitalization among children younger than 5 years) (16, 17). In this study, influenza viruses (IFVA-H1N1 + IFVA-H3N2+IFVB) (1253/6043, 20.73%) were the second most commonly detected RPs in the winter.
In summary, HRV and MP were the most commonly detected pathogens among children with ARTI in Henan province. The positive rate of CH was independent of the season, while it was not true for the positive rate of other pathogens. The positive rate of influenza A (H1N1) was independent from age, while the positive rate of other pathogens was age-related. The multiplex-PCR assay, based on the 3500DX Genetic Analyzer platform, can quickly detect the RPs that are associated with increased risk of ARTI. This ability not only helps clinicians to make an accurate diagnosis but also promotes rational use of antibiotics. This study demonstrated species-level information on the pathogens, which can improve prevention and therapy in hospitalized children with ARTI.
It is necessary to mention some limitations of our study, including following a single-center design, which probably has affected the representativeness of our samples. Also, the study lasted only for one year, which may not fully capture the epidemic trends of RPs.