1. Background
Autoimmune hepatitis (AIH) represents one of the chronic inflammatory hepatopathies characterized by hepatocyte autoimmune responses, which may lead to cirrhosis and liver failure (1). Autoimmune hepatitis occurs worldwide, and a national epidemiological survey in Finland suggested the AIH prevalence to be 14.3/100,000 in 2019 (2). In the United States, the prevalence of AIH was approximately 31.2 per 100,000 from 2014 to 2019 (3).
Hereditary spherocytosis (HS), a frequently observed familial hemolytic disease, results from erythrocyte membrane protein-encoding gene mutations, most of which are autosomal dominant, and a few of which are autosomal recessive (4). The typical clinical symptoms of patients with HS are anemia, jaundice, and splenomegaly (5). Like AIH, HS is observed globally. In the Nordic region, the incidence of HS was as high as 1/2000 (6), and its prevalence in China was about 1 per 100,000 persons (7).
To date, the pathogenic variants of the genes SPTB, ANK1, SLC4A1, EPB42, and spectrin alpha, erythrocytic 1 (SPTA1), which encode the erythrocyte membrane proteins β-spectrin, ankyrin, band 3 protein, 4.2 protein, and α-spectrin, respectively, have been associated with HS (8). Patients with both HS and AIH have not been reported in China.
2. Objectives
This study describes a seven-year-old girl with both HS and AIH, who was admitted to the First Affiliated Hospital of the Guangxi Medical University. To improve the clinical understanding of this rare condition, we analyzed the clinical characteristics of HS and AIH in the patient and genetically analyzed the patient, her parents, and her paternal grandmother.
3. Methods
3.1. Clinical History
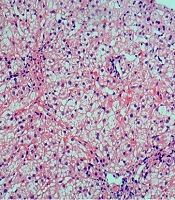
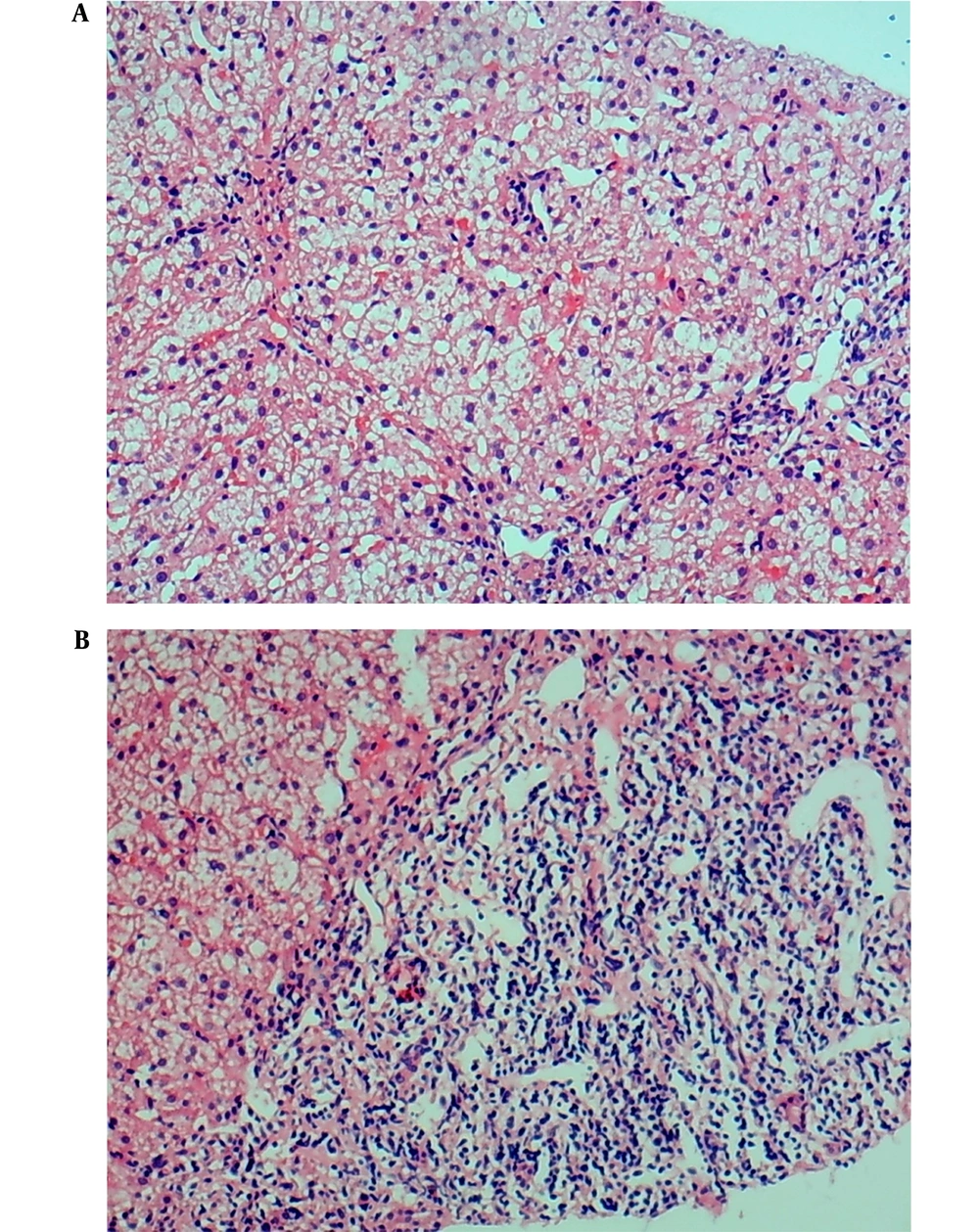
In June 2014, the patient was a girl born in the Guangxi Zhuang Autonomous Region, China. The four-year-old patient was admitted to our hospital with sclera yellowing for more than four months and liver function damage for four days. In April 2017, during her physical examination for kindergarten admission, the child was found with yellowish sclerae and slightly increased levels of transaminases and bilirubin. The yellow tinge of her sclerae disappeared spontaneously. Four months later, the child developed yellowish sclerae once more, and her transaminase and bilirubin serum levels were three times higher than the normal range. She was diagnosed with liver damage in another hospital and received liver protection treatment for two days. The yellowish sclerae was resolved, and she was further diagnosed and treated at our hospital. Health checkups exhibited yellowish sclerae, no obvious anemia, no lymph-node enlargement, and normal auscultation of both lungs. However, her liver and spleen were palpable at 4 cm under the xiphoid process and at 3 cm caudally of the lower ribs, respectively. Laboratory test results showed hemoglobin (Hb) of 114.7 g/L, reticulocyte percentage (RET) of 10.3%, red blood cell count (RBC) of 3.93 × 1012/L, mean corpuscular hemoglobin concentration (MCHC) of 359.10 g/L, mean sphered corpuscular volume (MSCV) of 60.29 fL, mean reticulocyte volume (MRV) of 73.04 fL, and mean corpuscular volume (MCV) of 81.35 fL. Spherocytes accounted for 18 % of the red blood cells (RBCs) in peripheral blood smears. The thalassemia gene test results showed no abnormality, glucose-6-phosphate dehydrogenase (G6PD) activity was normal, and the Coombs test was negative. Liver function tests revealed the following serum levels: total bilirubin (TBIL) of 64.30 μmol/L, indirect bilirubin (IBIL) of 32.30 μmol/L, direct bilirubin (DBIL) of 32.00 μmol/L, aspartate aminotransferase (AST) of 107 U/L, and alanine aminotransferase (ALT) of 137 U/L (Table 1). Investigating autoantibodies associated with the autoimmune liver disease showed positive antinuclear antibodies (ANAs) results. Immunofluorescence staining demonstrated homogeneous ANA karyotype (1: 100). Anti-smooth muscle antibodies (SMAs) were positive with a titer of 1: 320. Serum protein electrophoresis showed increased γ-globulins, and immunoglobulin (Ig)G was elevated by 28.89 g/L. All tests for hepatotropic viruses were negative, and the levels of blood copper, ceruloplasmin, and urine copper were normal. The histological examination of the liver biopsy showed diffuse swelling of cells in the liver lobules, cytoplasmic loosening from the cell wall, and spotty necrosis; however, no piecemeal or bridging necrosis was noticed. Although extensive lymphocyte infiltration in the portal area was observed, no obvious fibrous tissue hyperplasia was detected (Figure 1).
| Variables | Proband | Paternal Grandmother | Father | Mother | Reference Range |
|---|---|---|---|---|---|
| RBC (× 1012/L) | 3.93 | 3.61 | 4.81 | 4.71 | 3.50 - 5.50 |
| Hb (g/L) | 114.70 | 92.30 | 128.20 | 138.40 | 110.00 - 160.00 |
| MCV (fL) | 81.35 | 72.45 | 76.61 | 88.39 | 82.00 - 100.00 |
| MCH (pg) | 29.22 | 25.57 | 26.65 | 29.41 | 27.00 - 34.00 |
| MCHC (g/L) | 359.10 | 352.90 | 347.90 | 332.80 | 316.00 - 354.00 |
| MSCV (fL) | 60.29 | 83.80 | 87.80 | 84.08 | 84.00 - 104.00 |
| MRV (fL) | 73.04 | 82.95 | 90.50 | 99.05 | 101.00 - 119.00 |
| RET (%) | 10.30 | 2.40 | 2.30 | 1.00 | 0.00 - 2.00 |
| TBIL (μmol/L) | 64.30 | 23.60 | 16.70 | 8.50 | 3.40 - 20.50 |
| DBIL (μmol/L) | 32.00 | 3.80 | 2.80 | 2.50 | 0.00 - 6.80 |
| IBIL (μmol/L) | 32.30 | 19.80 | 13.90 | 6.00 | 3.10 - 14.30 |
| ALT (U/L) | 137 | 32 | 28 | 19 | 7 - 45 |
| AST (U/L) | 107 | 37 | 33 | 26 | 13 - 40 |
| SF (ng/mL) | 131.52 | 150.14 | 165.40 | 174.04 | 4.63 - 204.00 |
| SI (μmol/L) | 16.10 | 18.90 | 21.30 | 24.20 | 9.00 - 27.00 |
| TIBC (μmol/L) | 55.06 | 64.80 | 62.17 | 68.24 | 54.00 - 77.00 |
| UIBC (μmol/L) | 38.96 | 45.90 | 40.87 | 44.04 | 38.00 - 64.00 |
| ISAT (%) | 29.24 | 29.17 | 34.26 | 35.46 | 25.00 - 45.00 |
Abbreviations: RBC, red blood cell count; Hb, hemoglobin; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; MSCV, mean sphered corpuscular volume; MRV, mean reticulocyte volume; RET, reticulocyte ratio; TBIL, total bilirubin; DBIL, direct bilirubin; IBIL, indirect bilirubin; ALT, alanine aminotransferase; AST, aspartate aminotransferase; SF, serum ferritin; SI, serum iron; TIBC, total iron binding capacity; UIBC, unsaturated iron binding capacity; ISAT, iron saturation.
Histological investigation of liver biopsy in a pediatric patient with autoimmune hepatitis and hereditary spherocytosis (hematoxylin-eosin staining, × 100). A, Diffuse swelling of liver cells in the liver lobule and cytoplasmic loosening are visible; B, extensive lymphocyte infiltration in portal area.
3.2. Pedigree Investigation
The marriage of the proband’s parents was not consanguineous. Her father, mother, and paternal grandmother exhibited no clinical manifestation. We decided to further perform laboratory tests and the HS gene mutation analysis on the proband and her parents and paternal grandmother to identify the etiology of impaired liver function further.
3.3. Sample Collection
The proband’s parents signed the informed consent form to conduct each procedure of the study. The study protocols were approved by the Ethics Committee of the First Affiliated Hospital of the Guangxi Medical University. All procedures were carried out in line with the Declaration of Helsinki. In this study, we drew peripheral venous blood from the proband and her family members using three tubes per person. Two tubes were used to collect EDTA-K2-anticoagulated whole blood, one of which was used for routine blood tests, reticulocyte detection, and peripheral red blood cell morphology examination. Red blood cell parameters were analyzed with a Coulter LH780 Hematology Analyzer (Beckman Coulter, Brea, CA, USA). The second tube was used to extract genomic DNA from peripheral blood with a DNA extraction kit (Beijing Tiangen Biochemical Technology Co., Ltd., Beijing, China). The third tube contained separation gel for blood coagulation and was used for liver function tests with a 7600 Automatic Biochemical Analyzer (Hitachi Ltd., Tokyo, Japan).
3.4. Erythrocyte Osmotic Fragility Test
The erythrocyte osmotic fragility test was carried out as described in previous studies (9). In short, EDTA blood samples were added in a series of saline at gradient contents (NaCl, 0.0 - 0.9% w/v), followed by 120-min incubation under ambient temperature. Then the samples were centrifuged for 5 minutes at 200 g, and the results were determined by naked eye visualization result determination under naked-eye visualization. Reddish-clear supernatant was observed in hemolysis positive tubes, while colorless transparent supernatant was in hemolysis negative tubes. The initial and complete hemolysis values of the proband and her family members were recorded.
3.5. Next-Generation Sequencing
The extracted genomic DNA of the proband was sent to the Jinzhun Medical Laboratory (Beijing Jinzhun Gene Technology Co., Ltd., Beijing, China). The sequence variations of the whole exon coding regions and splicing regions of blood disease genes were analyzed by target region capture and the high-throughput sequencing technology (Illumina HiSeq2500 System, Illumina, San Diego, CA, USA). We utilized Burrows-Wheeler Aligner to align sequences and then detected mutations with the Genome Analysis Toolkit (Broad Institute, Cambridge, MA, USA; https://gatk.broadinstitute.org/hc/en-us) and VarScan (Genome Institute, Washington University, Washington, WA, USA; http://varscan.sourceforge.net/) software for the detection, annotation, and statistical analysis of single-nucleotide polymorphisms (SNP), InDels, and the others. Information was obtained from an external database using ANNOVAR software (https://annovar.openbioinformatics.org/) to evaluate the effects of a given sequence mutation and list the mutations (10). According to the sequencing analysis results, HS-related gene mutations were first considered, and the pathogenic mutations were detected by referring to the dbSNP, 1000G, and EXAC databases.
3.6. Sanger Sequencing
According to the next-generation sequencing results, we detected and verified mutations in the proband, her parents, and her paternal grandmother using PCR-Sanger sequencing. NM_003126 served as the reference transcript of SPTA1. This study utilized the following primers for PCR: SPTA1 c.134G>A (p.R45K) forward 5'-ACTTCCCACTCCACTGACT-3' and reverse 5'-TGACACATATAAGCGGGGCA-3'; and SPTA1 c.6544G>C (p.D2182H) forward 5'-GCAATGTAGGCAAGATTCCGT-3' and reverse 5'-TCTCAGAGTGGGAGAGGCC-3'. The PCR test was performed with I-5TM 2 × High-Fidelity Master Mix (MACLAB, San Francisco, CA, USA) using a 50 µL solution containing 2 × Taq PCR Master Mix (25 µL, Sangon Biotech Co., Ltd., Shanghai, China), water (21 µL), cDNA (2 µL), and respective primers (1 µL). The PCR procedure was conducted as follows: 5-min initial denaturation < 95°C; 30-s < 95°C, 30-s < 60°C, and 40-s < 72°C for 35 cycles, and 8-min eventual extension < 72°C.
3.7. Pathogenicity Prediction and Analysis of Gene Mutation Conservation
For the SPTA1 gene’s mutation sites c.134G>A (p.R45K) and c.6544G>C (p.D2182H), Mutation Taster (http://www.mutationtaster.org/), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/index.shtml), PROVEAN (http://provean.jcvi.org/index.php, J. Craig Venter Institute, La Jolla, CA, USA), and SIFT (https://sift.bii.a-star.edu.sg/) were used to evaluate their pathogenicity (11-13). We also utilized DNAMAN (Lynnon Biosoft, San Ramon, CA, USA) to analyze the conserved amino acid sequence of SPTA1 among different species.
3.8. Protein Structure Analysis
HOPE software (https://www3.cmbi.umcn.nl/hope/) (14) was used to conduct the homologous modeling of the SPTA1 gene’s mutation sites c.134G>A (p.R45K) and c.6544G>C (p.D2182H) to predict the effect of amino acid residue variations caused by gene mutations affecting protein functions. We obtained the sequences according to SPTA1 in UniProtKB (https://www.uniprot.org/uniprot/P02549).
4. Results
4.1. Laboratory Results of Proband and her Family Members
Table 1 displays the routine blood and reticulocyte test results for the proband and her family members. Table 2 shows the additional laboratory test results for the proband.
| Variables | Results | Reference Range |
|---|---|---|
| IgG (g/L) | 28.890 | 8.00 - 18.00 |
| IgA (g/L) | 0.840 | 0.90 - 4.50 |
| IgM (g/L) | 2.230 | 0.84 - 1.32 |
| γ-globulin | 26.00 | 11.10 - 18.80 |
| ANA | + | - |
| SMA | + (Titer 1: 320) | - |
| ANA karyotype | Homogeneous (1: 100) | - |
| LKM-1 | - | - |
| LC-1 | - | - |
| AFP (ng/mL) | 84.73 | 0.89 - 8.78 |
| HBS Ag | 0.00 | 0.00 - 0.50 |
| HAV-IgM | - | - |
| HCV-Ab | - | - |
| HEV-Ab | - | - |
Abbreviations: -, negative; +, positive; Ig, immunoglobulin; ANA, antinuclear antibody; SMA, smooth muscle antibody; LKM-1, anti-liver and kidney microsomal antibody type 1; LC-1, antibody against liver cytosol type-1 antigen; AFP, alpha-fetoprotein; HBS Ag, hepatitis B surface antigen; HAV-IgM, hepatitis A IgM antibody; HCV-Ab, hepatitis C antibody; HEV-Ab, hepatitis E antibody.
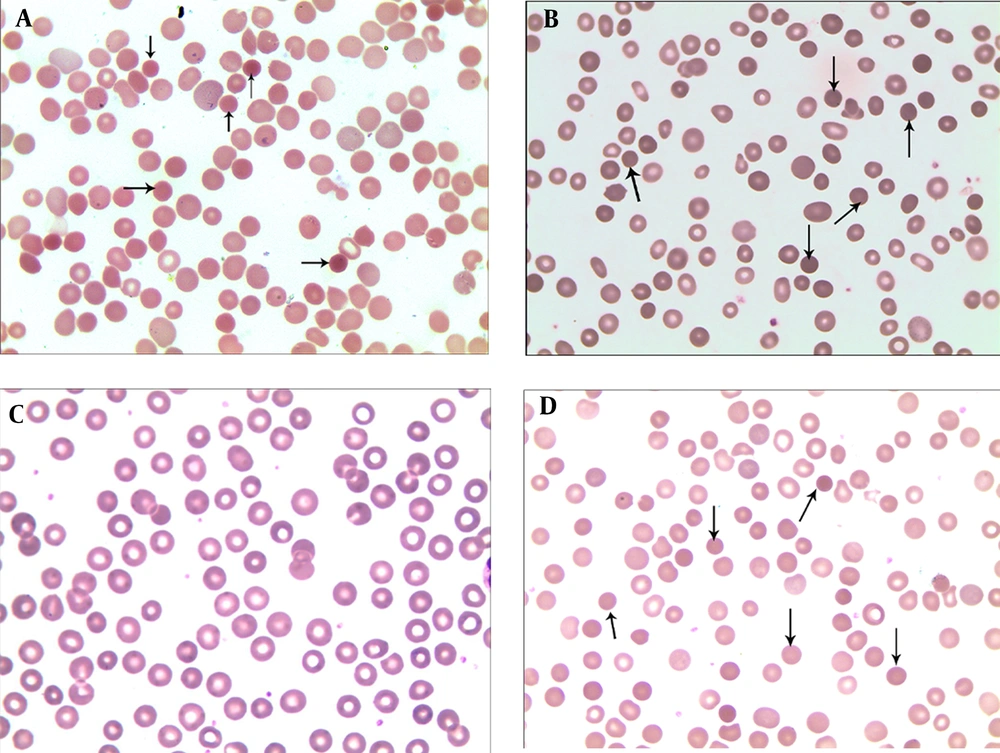
The proband’s peripheral blood smears revealed that spherical cells that accounted for 18% of the red blood cells. Spherical cells accounted for 10% and 12% of the red blood cells in the peripheral blood smears of the father and paternal grandmother, respectively. The mother’s peripheral blood smear results were normal (Figure 2).
4.2. Erythrocyte Osmotic Fragility Test
The erythrocyte osmotic fragility test results determined the values of complete (normal range: 3.20 - 3.40 g/L) and initial (4.2 - 4.6 g/L) hemolysis of 5.30 and 6.0 g/L for the proband, 3.3 and 4.40 g/L for the proband’s father, 3.1 and 4.70 g/L for the proband’s paternal grandmother, and 3.3 and 4.30 g/L for the proband’s mother. Other laboratory findings for the proband and her family members showed no abnormality in the thalassemia gene test, normal G6PD activities, and negative Coombs tests.
4.3. Genetic Test Results for Hereditary Spherocytosis
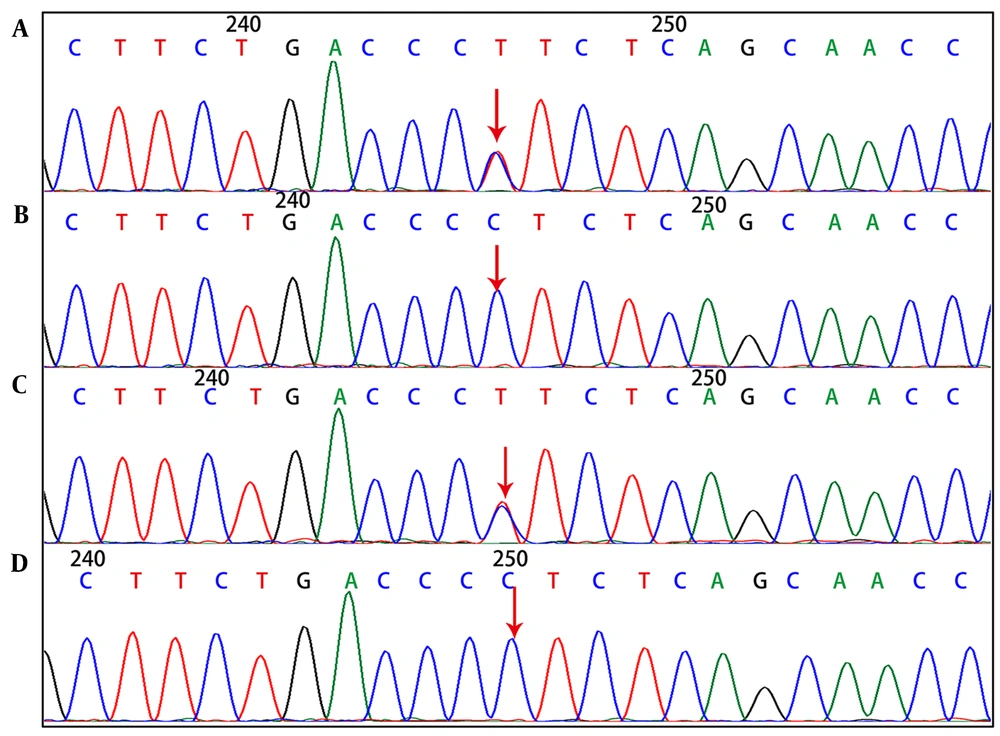
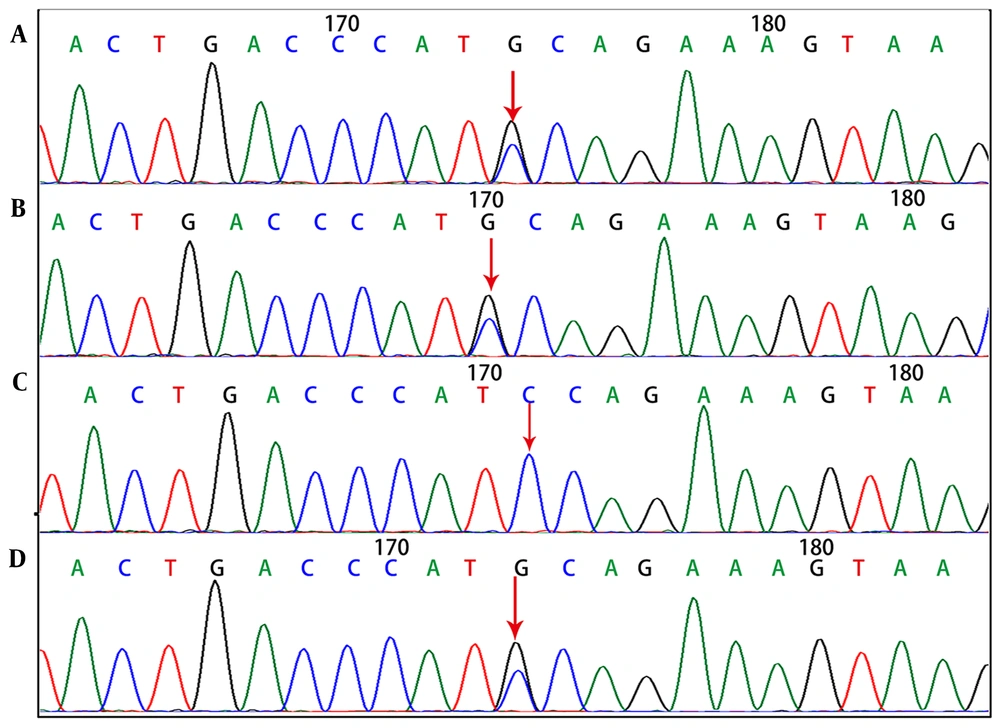
Genetic tests showed that the SPTA1 gene of the proband, had novel exon 46 c.6544G>C (p.D2182H) and exon 2 c.134G>A (p.R45K) compound heterozygous mutations. The c.134G>A (p.R45K) mutation of SPTA1 was inherited from the mother (Figure 3), whereas the c.6544G>C (p.D2182H) mutation of SPTA1 was inherited from the father.
In this regard, the paternal grandmother had the same mutation as the father (Figure 4).
Findings of SPTA1 gene c.6544G>C (p.D2182H) mutation. A, Heterozygous mutation for proband; B, heterozygous mutation for father; C, normal for mother; D, heterozygous mutation for grandmother (the arrows in figures A, B, and D indicate the mutation sites, while the arrows in figure C show no mutation).
4.4. Prediction of Gene Mutation Pathogenicity and Conservation Analysis
SPTA1 c.134G>A (p.R45K) is a novel missense mutation not detected from the Human Gene Mutation Database. No pathogenicity analysis of this site was included in the ClinVar database; however, the mutation was predicted to be ‘benign’ in Polyphen-2 (score 0.052), ‘disease-causing’ in Mutation Taster (26), ‘tolerated’ in SIFT (0.478), and ‘neutral’ in Protein Variation Effect Analyzer (PROVEAN).
SPTA1 c.6544G>C (p.D2182H) is also a novel missense mutation not discovered in Human Gene Mutation Database. No pathogenicity analysis of this site was included in the ClinVar database. Polyphen-2 (score 0.990), Mutation Taster (81), SIFT (0.001), and PROVEAN (-3.09) predicted a pathogenic effect, described as ‘probably damaging’, ‘disease-causing’, ‘damaging’, and ‘deleterious’, respectively. The conservative sequence analysis showed that the c.134G>A (p.R45K) and c.6544G>C (p.D2182H) sites of SPTA1 were highly conserved among different species (Figure 5). According to the guidelines of the American College of Medical Genetics and Genomics (ACMG), the variants p.R45K and p.D2182H are the variants of uncertain significance (VUS).
4.5. Protein Structure Analysis of SPTA1 c.134G>A Mutation
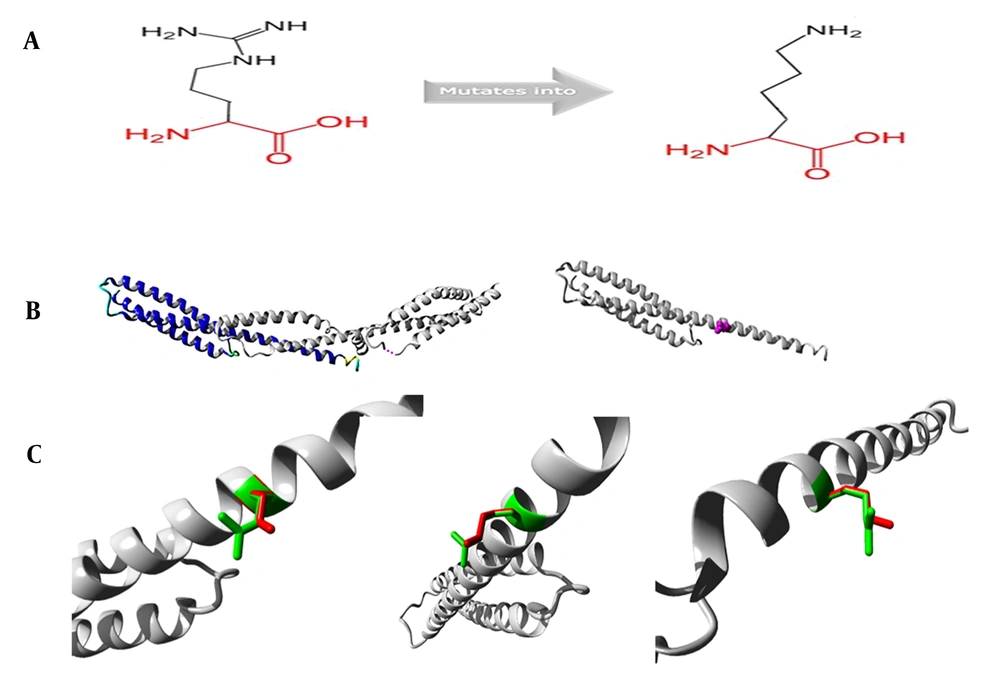
The mutation SPTA1 c.134G>A (p.R45K) was analyzed for its protein structure; therefore, mutant residue decreased compared to wild-type (WT) one. Moreover, WT residue and aspartic acid formed the hydrogen bond at the position 291. Differences in size indicates the incorrect position of mutant residue in forming hydrogen identical to the initial WT one (Figure 6). Schematics for c.6544G>C (p.D2182H) were not prepared due to the unavailability of 3D structures from the HOPE database (14).
SPTA1 c.134G>A (p.R45K) analysis for its protein structure. A, Sketch maps illustrates the original (left) and mutant (right) amino acid structures (the backbone is red, and it is identical to all amino acids; the side chain specific for a diverse amino acid is black). B, The left panel displays a protein overview in terms of ribbon presentation (the following elements denote protein: α-helix, blue; turn, green; β-strand, red; random coil, cyan, and 3/10 helix, yellow). Additional molecules within the complex are gray if existed. The right panel displays the mutant protein overview in terms of ribbon presentation. Protein is gray, while the side chain specific for mutant residue is displayed in small balls and denoted in magenta. C, Three mutant structure angles. Protein is gray, while side chains specific for mutant and WT residues are denoted in red and green, respectively.
5. Discussion
Autoimmune hepatitis is an inflammation of the liver parenchyma caused by an autoimmune response to hepatocytes, which is characterized by elevated transaminase serum levels, positive serum autoantibodies, high IgG and/or γ-globulinemia, and interstitial hepatitis upon histological examination (1). Regarding autoantibody patterns, AIH is further classified into three subtypes: AIH-1, AIH-2, and AIH-3. AIH-1 is characterized by SMA, ANA, or both. AIH-2 is characterized by the presence of specific anti-kidney and anti-liver microsomal antibody type-1, whereas anti-liver cytosol type-1 and anti-LKM type-3 antigen antibodies are rare. AIH-3 is characterized by soluble liver antigen/liver pancreas antibodies (15). The clinical manifestations of AIH are different; however, most patients have an insidious onset represented as chronic liver diseases (16). The most common symptoms include lethargy, fatigue, and intermittent jaundice; however, physical examination may reveal hepatomegaly, splenomegaly, ascites, and, occasionally, peripheral edema (1, 17). In our study, the patient presented with yellow sclerae, liver and spleen enlargement; elevated levels of ALT, AST, TBIL, IBIL, and DBIL, positive ANA and SMA, slightly increased IgG, and increased γ-globulin levels. Her liver biopsy revealed diffuse swelling of the liver lobule cells, cytoplasmic loosening, spotty necrosis, and extensive lymphocyte infiltration in the portal area. According to the findings, the patient was diagnosed with AIH. As the morphological examination of her peripheral red blood cells indicated, 18% of the cells were spherical, the MCV, MSCV, and MRV levels were decreased, RET was increased; G6PD activity was normal, and the Coombs test was negative. These findings suggested that the patient's abnormal liver function was not the result of AIH alone and that she might have also been suffering from hemolytic anemia.
Hereditary spherocytosis is a common hereditary hemolytic disease. Gene mutations lead to defects in erythrocyte membrane proteins, which decrease the longitudinal associations of a lipid bilayer with protein (18), cause membrane lipid bilayer loss or destabilization and the formation of microvesicles, reduce the cell membrane surface area to volume ratio, and change the red blood cell’s morphology from a biconcave disc to a spherical shape. These changes reduce deformability and increase the osmotic fragility of erythrocytes, resulting in their destruction when passing through the small splenic vessels and, ultimately, hemolytic anemia. Clinical manifestations vary widely among HS patients. Patients with severe and moderate HS often have anemia, jaundice, and splenomegaly, whereas those with mild HS have either atypical symptoms or are asymptomatic (4). As suggested by the British Committee for Standards in hematology (6) guidelines on the diagnosis and treatment of HS, patients with a positive family history, typical clinical features, and increased MCHC, spherocytosis, and reticulocytes, compared to normal values, are diagnosed with HS without further examination. Accordingly, patients with mild HS and no symptoms are either easily misdiagnosed, or the diagnosis is entirely missed. Furthermore, because of the similar clinical manifestations of HS and AIH, such as jaundice and splenomegaly, the HS diagnosis is frequently missed even in symptomatic patients, and it is misdiagnosed as AIH.
Deng et al. (19) reported a patient with mild HS who had jaundice but no symptoms of anemia and was misdiagnosed with AIH. The type of jaundice can be classified by measuring TBIL, IBIL, and DBIL. Cases with hepatobiliary diseases usually exhibit higher TBIL, mainly attributable to increased DBIL. Patients with hemolytic anemia also have increased TBIL, which can primarily be attributed to increased IBIL. In our patient, TBIL was increased because of both DBIL and IBIL, with each factor accounting for 50%. The patient had hepatomegaly and splenomegaly but no anemia symptoms, which might have easily led to the misdiagnosis of HS.
HS can be rapidly diagnosed with laboratory blood count, erythrocyte morphology, or the assessment of reticulocyte-associated variables. If spherocytes made up above 7.8% of RBCs, the specificity and sensitivity in the diagnosis of HS would be 84.8% and 56.7%, respectively (20). Moreover, spherocytes may exist within peripheral blood smears from cases with G6PD deficiency and autoimmune hemolytic anemia (5). Liao et al. (21) reported that, with MCHC ≥ 334.9 g/L as the threshold, the HS diagnosis sensitivity and specificity were 82.1 and 94.5%, respectively. The elevated bilirubin level in serum may cause a false increase in MCHC. MSCV represents a unique measuring method for spherocytes, and when it is used with MCV, the efficiency of the HS diagnosis is greatly enhanced. As Chiron et al. suggested (22), for the MSCV < MCV, the sensitivity, and specificity of the HS diagnosis were 100 and 93.3%, respectively. According to Broseus et al. (23), when the Coombs test was negative and MCV-MSCV > 9.6 fL, the specificity and sensitivity of the HS diagnosis were 90.57 and 100%, respectively. In another study, Xu et al. (24) reported that MRV was ideal for the HS diagnosis, with the specificity and sensitivity of 91.20 and 86.80% at MRV ≤ 95.77 fL, respectively. Our patient met all diagnostic criteria; hence, her laboratory results strongly indicated HS; however, she had no typical clinical symptoms or positive family history. As a consequence, the genetic analysis became necessary for an accurate diagnosis. Using high-throughput sequencing, we found two novel compound heterozygous mutations within exon 46 c.6544G>C (p.D2182H) and exon 2 c.134G>A (p.R45K) from SPTA1. PCR combined with Sanger sequencing confirmed that the first mutation was from the asymptomatic mother, and the second mutation was from the father.
About 75% of the HS cases result from an autosomal dominant inheritance, which mostly includes ANK1, SPTB, or SLC4A1 gene mutations. However, the other 25% are related to novel mutations or autosomal recessive inheritance. In this regard, SPTA1 and EBP42 are the most frequently detected compound heterozygous mutations (25). Being located within the chromosome 1 q22-q23 region, the 80 Kb SPTA1 gene possesses 52 exons, contains 2419 amino acids, and can encode α-spectrin, which is a cytoskeletal protein combined with β-spectrin to form α-β heterodimers in a reverse parallel arrangement. The terminal end of the polymers formed by the two heterodimers combines with actin and transmembrane proteins forming a "junction complex." The latter forms a grid structure in the cell membrane, thereby maintaining lipid bilayers and the biconcave disc shape of the erythrocytes (26). The expression of α-spectrin is four times higher than that of β-spectrin, and the formation of α-β heterodimers is not affected by relative decreases in the α-spectrin expression (27). Consequently, compound heterozygous or homozygous mutations within SPTA1 are possibly related to the pathogenic mechanism of HS with deficient recessive α-spectrin. Individuals with heterozygous mutation are likely to generate sufficient α-spectrin to balance β-spectrin synthesis while maintaining cytoskeleton of erythrocytes (28, 29). The SPTA1 missense mutation of c.134G>A (p.R45K) made the 45th amino acid change from arginine to lysine. The amino acid sequence analysis showed that the mutation site was highly conserved among different species. In the protein structure analysis, mutant residue had a decreased size compared to WT one, while the latter was associated with the multimer contact. Mutant residue may be too small to make multimer contacts, thereby negatively affecting the function of α-spectrin. The missense mutation of c.6544G>C (p.D2182H) made the 2182nd amino acid change from aspartic acid to histidine. This site is also highly conserved among different species. The protein structure analyses suggested that the mutation was located in the protein residue repeated stretch, referred to as spectrin 20. Moreover, another residue mutation probably broke such a repeat or the associated functions. The mutant residue lies within the domain required for binding additional molecules, which may thus disturb this function (14) Furthermore, we speculated that c.6544G>C (p.D2182H) mutation played the pathogenic role via Mutation Taster, Polyphen-2, PROVEAN, and SIFT. Accordingly, the heterozygous mutations of the novel compound c.134G>A (p.R45K) and c.6544G>C (p.D2182H) in the SPTA1 gene might have caused HS in the patient with AIH.
5.1. Conclusion
Patients with both HS and AIH are rare. HS displays clinical presentations close to AIH; therefore, it is challenging to diagnose HS in the co-existing of AIH. If a single autoimmune hepatopathy cannot explain laboratory results, there may be a concurrent disorder. In this case, the lesion should be diagnosed by genetic analyses and pedigree investigations