1. Background
As one of the highly prevalent diseases and a syndrome with a lethal outcome, neonatal sepsis is always accompanied by hemodynamic changes and other systemic clinical manifestations directly caused by pathogenic microorganisms. It is also the main cause of neurocognitive sequence and mortality (1, 2). Neonatal sepsis is identified as a syndrome in the first month of patients' lives. According to the time of onset, it is divided into early or late neonatal sepsis. The early one is considered when the clinical condition appears within the first 72 h of life, while the late neonatal sepsis starts after 72 h of life (1, 2). Current knowledge suggests that bacteria, viruses, and fungi, which are normally present in sterile fluid, are the main causes of neonatal sepsis, greatly facilitating diagnosis (3). However, reaching an accurate diagnosis is always challenging in clinical settings. Unlike adult patients, newborns have very subtle presentations and multiple influences (4). Due to the low sensitivity and/or specificity of clinical parameters and blood culture results, coupled with the age-specific medical reference ranges, the diagnosis of neonatal diseases is more complex and difficult (5). Moreover, the early-onset neonatal sepsis is occurring within 72 hours of birth. It results from organisms acquired intrapartum and might be associated with maternal genitourinary tract status, with infections of group B streptococcus and gram-negative enteric bacteria. Late neonatal sepsis often occurs in hospitalized infants and/or undergoes invasive procedures; it is usually associated with the presence of common microorganisms in the hospital settings (6-8).
2. Objectives
Thus, in this study, the authors aim to use common clinical characteristics/parameters to develop diagnostic models for distinguishing bacterial with/without viral infections among neonatal sepsis patients and to evaluate the prognostic value of the models.
3. Methods
3.1. Patients
The authors reviewed the records of Tianjin Children's Hospital from 2017 to 2019 and then followed them up from 2019 to 2021. A total of 479 patients were included in this study. Blood cultures were used to confirm sepsis, with a complete profile and information needed for the analysis. In this study, the cases between 2017 and 2019 (n = 368) were used as the training cohort, and the cases from 2019 to 2021 were used as the verifying cohort (n = 111). Inclusion criteria included the early sepsis patients with complete clinical and laboratory data. Sepsis was suspected in the presence of unstable temperature, feeding intolerance, respiratory distress, lethargy, hemodynamic instability, convulsion, hypotonia, or irritability. Then, sepsis was identified by positive blood culture. Exclusion criteria included individuals with severe congenital malformations, asphyxia, liver disease, renal failure, malignant tumor, drug poisoning, diabetes, a history of genetic metabolic diseases or familial immune diseases, and incomplete medical records. The institutional review board of the hospital waived the requirement for patients' informed consent.
3.2. Clinical Parameters
Relative information was collected from the hospital profile database. Clinical features and related clinical parameters and biochemical indicators included 16 factors such as gender, age, infant age, birth weight, maternal infection, cough, diarrhea, high fever, chills, moan, vomiting, higher level of IL-6, C-reactive protein (CRP), and hemoglobin (HGB), White Blood Cell (WBC) count, and platelet (PLT) level.
3.3. Statistical Analysis
The SPSS 19.0 software package was used for data analyses. The Mann-Whitney U test was conducted to evaluate the significance of clinical parameters. Backward elimination methods of the Logistic Regression model were used to identify the optimum feature combination by adding all potential factors to the regression equation. The potential predictors included in the univariate analysis were based on clinical knowledge and literature review (7, 8). The variables selected for multivariate analysis were based on univariate analysis, while the marginal variable inclusion was based on both univariate analysis and clinical knowledge. Furthermore, 10-fold cross-validation was carried out in order to evaluate the model fitting, and the accuracy of LOO-CV was calculated. Also, model performance was verified by the Receiver Operating Characteristic (ROC) curve and the Area Under the Curve (AUC), together with the accurate level of sensitivity and specificity. Also, P < 0.05 was considered significant.
4. Results
4.1. Comparability of the Training Cohort and Verifying Cohort
The current study established 3 predictive models: (1) Diagnosis model 1: To distinguish the bacterial infection and non-bacterial infection, 329 out of 368 cases were included; (2) diagnosis model 2: To distinguish the bacterial infection with/without virus infection, 265 out of 368 cases were included; (3) prognosis model: To predict the hospital stay (as one aspect of short-term prognosis) whether longer than 7 days, all the 368 cases were included.
The training cohort (3 sets of training cohort: Diagnosis model 1, diagnosis model 2, and diagnosis model 3) and verifying cohort were comparable, with no statistical difference in gender, age, gestational age, and birth weight distribution (Table 1). However, the distribution of hospital days indicated a statistical difference in model 2, which might be related to the specific disease or prognosis status.
| Cohort | Male:Female | Pre-term, No. (%) | Age, m | Gestational Age, d | Birth Weight, kg | Hospital Days |
|---|---|---|---|---|---|---|
| Training cohort 1 (n = 329) | 178:151 | 21 (6.38) | 14.220 ± 8.152 | 271.066 ± 15.860 | 3.258 ± 0.497 | 12.350 ± 9.591 |
| Verifying cohort 1 (n = 111) | 63:38 | 12 (10.81) | 14.511 ± 8.600 | 270.918 ± 12.054 | 3.286 ± 0.568 | 12.400 ± 10.186 |
| t(χ2)/P | 2.147/0.143 | 2.346/0.096 | 1.423/0.156 | 0.103/0.928 | 0.495/0.621 | 0.047/0.963 |
| Training cohort 2 (n = 265) | 30:23 | 25 (9.43) | 13.085 ± 8.661 | 271.264 ± 15.686 | 3.267 ± 0.493 | 10.230 ± 8.898 |
| Verifying cohort 2 (n = 111) | 63:38 | 12 (10.81) | 14.511 ± 8.600 | 270.918 ± 12.054 | 3.286 ± 0.568 | 12.400 ± 10.186 |
| t(χ2)/P | 1.002/0.317 | 0.167/0.406 | 1.355/0.176 | 0.231/0.835 | 0.326/0.745 | 2.066/0.040 |
| Training cohort 3 (n = 368) | 201:167 | 25 (7.29) | 13.567 ± 8.704 | 271.363 ± 15.379 | 3.268 ± 0.494 | 10.360 ± 8.779 |
| Verifying cohort 3 (n = 111) | 63:38 | 12 (10.81) | 14.511 ± 8.600 | 270.918 ± 12.054 | 3.286 ± 0.568 | 12.400 ± 10.186 |
| t(χ2)/P | 1.938/0.164 | 1.390/0.163 | 1.004/0.316 | 0.319/0.780 | 0.325/0.746 | -1.907/0.054 |
a Cohort 1: Distinguishing bacterial infection patients and non-sepsis controls; cohort 1: Distinguishing bacterial infection patients and bacterial + viral infection patients; cohort 3: Distinguishing hospital days longer than 7 days and shorter than 7 days
4.2. Establishment of the Diagnostic Models from the Training Data Sets
In light of the backward elimination analysis, 16 factors covered the common high-frequency symptoms, and routine biochemical indicators and clinical parameters were finally used for modeling.
In distinguishing bacterial sepsis patients and bacterial culture-negative patients, we found that Y = 1.930+0.105X1+0.891X2-1.389X3-0.774X4, which Y symbolizes the status of bacterial infectious sepsis (1 = no, 2 = yes), X1 is age increase, X2 is intra-amniotic infection (mother) (1 = no, 2 = yes), X3 is vomiting sign (1 = no, 2 = yes), and X4 is cough sign (1 = no, 2 = yes). The OR values for relative factors from multi-variate logistic regression analysis are shown in Table 2.
| Variables | ß | S.E | Wald | OR (95% CI) | P-Value |
|---|---|---|---|---|---|
| Model 1 | |||||
| Age increase | 0.105 | 0.018 | 32.611 | 1.111 (1.072~1.152) | <0.001 |
| Intra-amniotic infection (mother) | 0.891 | 0.420 | 4.485 | 2.436 (1.069~5.555) | 0.034 |
| Vomiting | -1.389 | 0.516 | 7.233 | 0.249 (0.091~0.686) | 0.007 |
| Cough | -0.774 | 0.381 | 4.122 | 0.461 (0.218~0.974) | 0.042 |
| Model 2 | |||||
| IL-6 increase | 1.568 | 0.314 | 24.911 | 4.797 (2.592~8.879) | <0.001 |
| CRP increase | 1.882 | 0.917 | 4.211 | 6.565 (1.088~39.616) | <0.001 |
| Age increase | -0.113 | 0.037 | 9.301 | 0.893 (0.830~0.960) | 0.002 |
| High fever | -2.214 | 0.905 | 5.985 | 0.109 (0.019~0.644) | 0.014 |
| Cyanotic | -2.255 | 0.635 | 12.616 | 0.105 (0.030~0.364) | <0.001 |
| HGB increase | -2.312 | 0.587 | 15.530 | 0.099 (0.031~0.313) | 0.040 |
| Model 3 | |||||
| Age increase | 0.073 | 0.016 | 21.086 | 1.076 (1.043~1.110) | <0.001 |
| Intra-amniotic infection (mother) | 1.963 | 0.427 | 21.150 | 7.118 (3.084~16.430) | <0.001 |
| IL-6 increase | 0.466 | 0.176 | 7.014 | 1.594 (1.129~2.250) | 0.008 |
| Convulsion with unconsciousness | -0.791 | 0.410 | 3.727 | 0.453(0.203~1.012) | 0.054 |
| Cough | -0.633 | 0.348 | 3.312 | 0.531(0.268~1.050) | 0.069 |
a Model 1: Distinguishing bacterial infection patients and non-sepsis controls; model 2: Distinguishing bacterial infection patients and bacterial+viral infection patients; model 3: Distinguishing hospital days longer than 7 days and shorter than 7 days.
Similarly, we found in distinguishing bacterial sepsis patients and bacterial/viral double-positive patients: Y = 2.918+1.568X1+1.882X2-0.113X3-2.214X4-2.255X5-2.312X6, which Y means the status of bacterial/viral double-positive status (1 = no, 2 = yes), X1 is IL-6 increase, X2 means CRP increase, X3 means age increase, X4 means high fever sign (1 = no, 2 = yes), X5 is cyanotic sign (1 = no, 2 = yes), and X6 is HGB increase. The ORs for relative factors can be found in Table 2. Finally, the current analysis suggested that for predicting hospital days as one of the prognosis aspects, Y = -1.993+0.073X1+1.963X2+0.466X3-0.791X4-0.633X5, where Y means worse prognosis which is hospital days longer than 7 days (1 = no, 2 = yes), X1 means age increase, X2 means intra-amniotic infection (mother) (1 = no, 2 = yes), X3 is IL-6 increase, X4 is convulsion with unconsciousness (1 = no, 2 = yes), and X5 is cough sign (1 = no, 2 = yes). The OR values can be found in Table 2.
Furthermore, the Hosmer-Lemeshow test for 3 models showed that the χ2/P values are as follows: χ2 = 7.466, P = 0.487; χ2 = 6.034, P = 0.643, χ2 = 9.629, and P = 0.292, suggesting the goodness of fit for logistic regression. The cross-validation result further suggested that 70.5% of cross-validated grouped cases were correctly classified, and 74.8% of original grouped cases were correctly classified in diagnosis model 1; they were 92.6% and 90.9% in model 2 and 69.6% and 72.3% in model 3, respectively, showing a good fitting status.
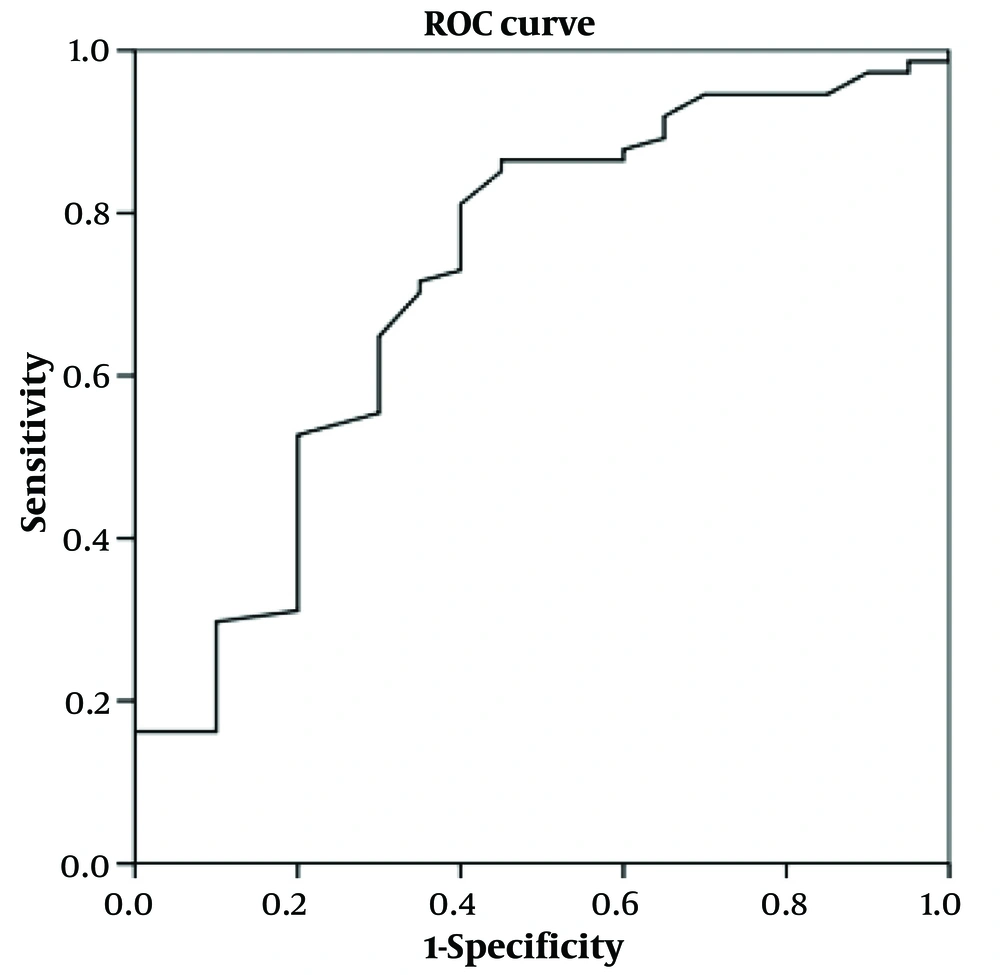
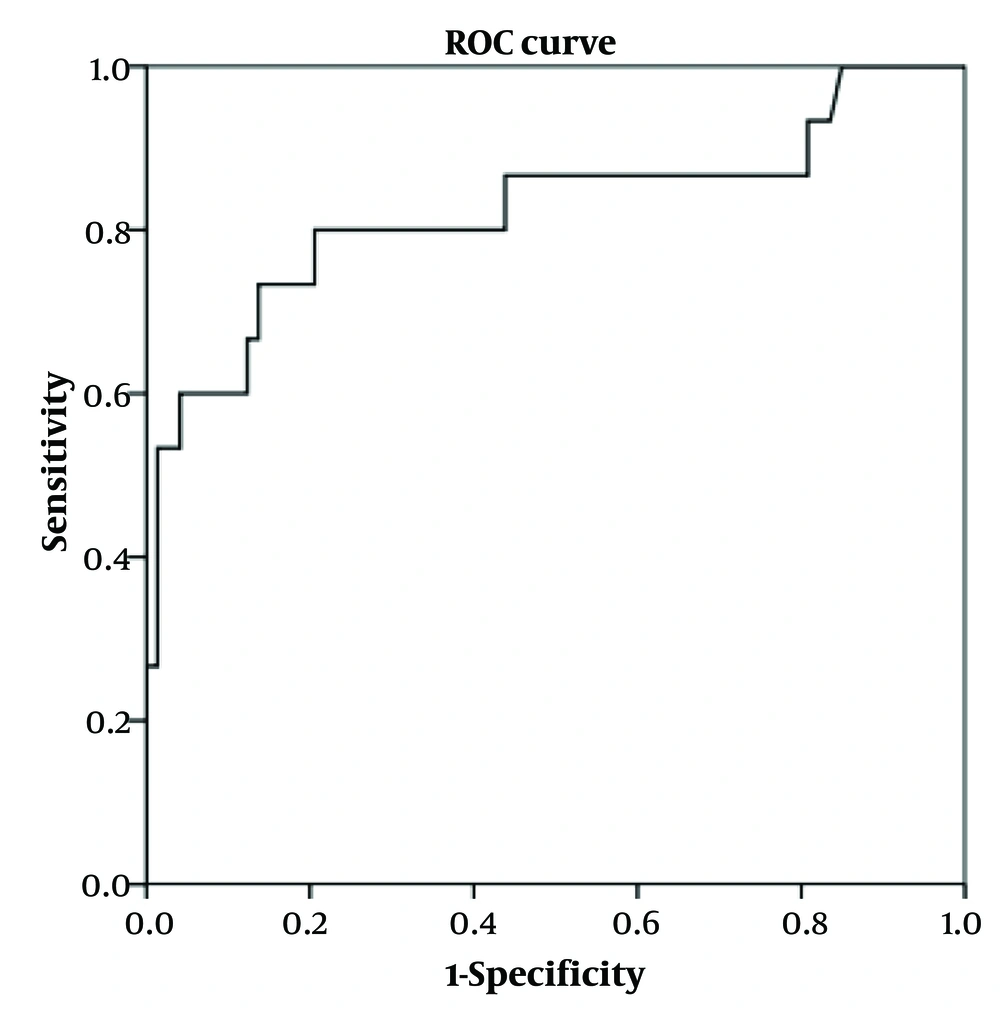
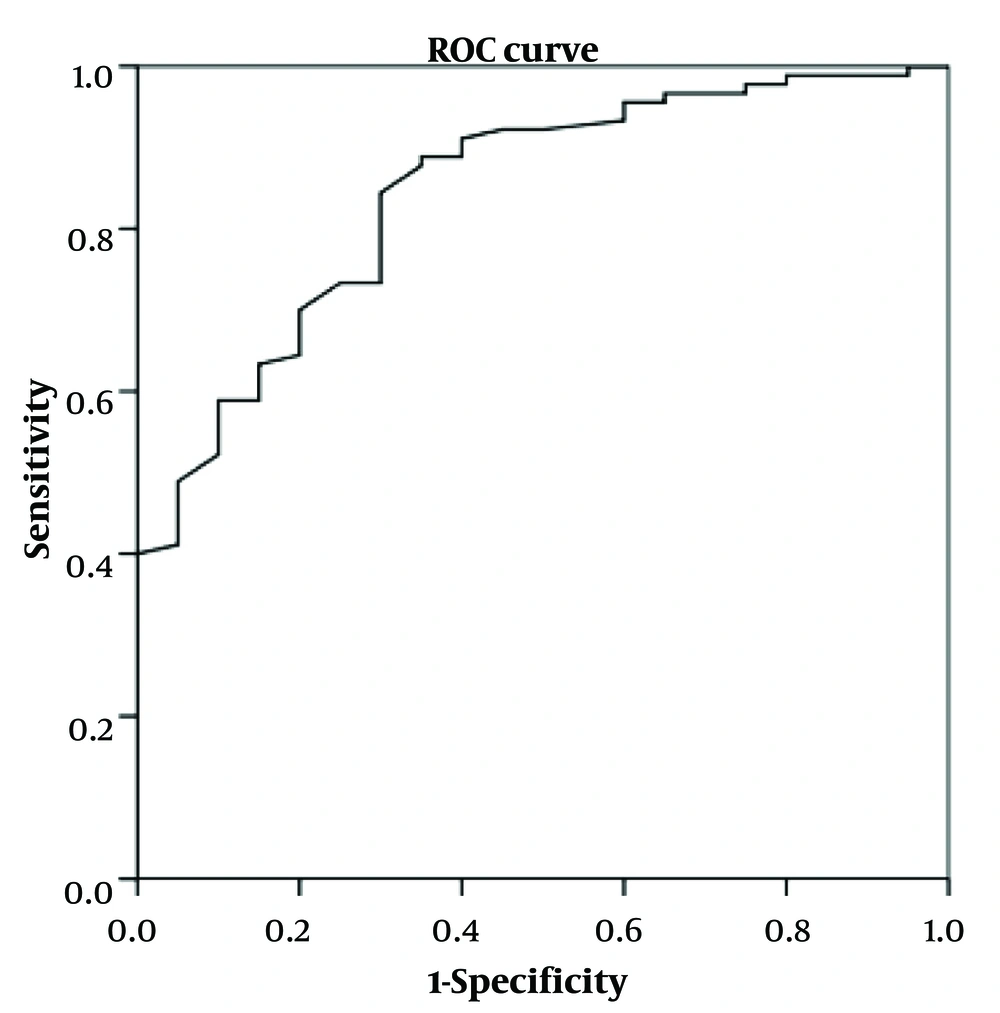
4.3. Validation of Models
The previous models were applied for testing using the verifying cohort. The ROC curves of the models in the verifying cohort are shown in Figures 1-3. The AUROC values were 0.721 (0.587 ~ 0.854, P = 0.003), 0.823 (0.678 ~ 0.968, P = 0.000), and 0.843 (0.755 ~ 0.932, P = 0.000) for models 1 to 3, respectively, indicating that the models had perfect performance. Thus, in further clinical work, the patients with y ≥ 0.856 (Se = 0.986, 1 - Sp = 0.950) have the best-predicting power for potential bacterial infection, as for the reference in treatment. Meanwhile, y ≥ -6.772 (Se = 0.933, 1 - Sp = 0.836) is the best-predicting power to predict bacterial/viral double-positive status. Moreover, in prognostic evaluation, children with y ≥ -0.608 (Se = 0.989, 1 - Sp = 0.950) have the best-predicting power for indicating longer hospital days.
5. Discussion
The World Health Organization acknowledges neonatal sepsis as a major global health concern and that the highest burden occurs in most of the developing areas (7). The etiological factors and symptoms of neonatal sepsis might be quite distinct (8, 9). The infections might be caused by different etiological agents (8-10). Moreover, different pathogens mean different diagnoses, treatments, and prognosis (10).
The clinical signs are also important for diagnosis and treatment. The common signs of sepsis can be divided into several groups: One group of signs is apnea and difficulty breathing, and the other group includes tachycardia or bradycardia, poor perfusion, or shock. The third group of common signs include irritability, lethargy, or hypotonia, while the fourth subgroup is jaundice. Also, the common signs include temperature instability, petechiae, or purpura (11). Till now, it is still not clear what the accurate signs are to differentiate between sepsis patients of viral origin and those of bacterial origin (11). The clinical experience shows that high fever and increased WBC count might be markers for bacterial infection, as disclosed decades ago (12).
As for those clinical parameters, there is currently a large effort to detect biomarkers that can aid clinicians in the diagnosis or evaluating the prognosis. The markers might be the different metabolites and proteins from human biological samples, which can offer predictive information. The expression of or modifications to them can provide a more reliable source of diagnostic or prognostic assessment, especially together with some of the special clinical parameters and indexes (13-15). Here, the authors collected and analyzed the common signs and biochemical indicators, including 16 factors: Gender, age, infant age, birth weight, maternal infection, cough, diarrhea, high fever, chills, moaning, and vomiting, while the potential associated clinical parameters were a higher level of IL-6, CRP, and HGB, WBC count, and PLT level. We found different predictive values for those factors when distinguishing pathogen origin and prognosis. Since the current diagnosis continues to rely primarily on inaccurate microbiologic techniques, the possible misdiagnosis and inappropriate treatment would lead to a worse prognosis accordingly. Although it is very important, no standard diagnostic model based on signs has been established for neonatal sepsis to date. Thus, a hospital-based sample was recruited to establish the predictive model for clinical reference.
In the current study, bacterial infectious sepsis might be mainly caused by age increase and intra-amniotic infection of the mother (y ≥ 0.856). In contrast, signs of vomiting and cough showed the opposite effect. To distinguish bacterial/viral double-positive patients, the alarmed factors included IL-6 increase and CRP increase, while age increase, high fever, cyanotic sign, and HGB increase were the negative indications. Similarly, we could assess the possible bacterial/viral double infection by checking y ≥ -6.772. Moreover, based on the current diagnostic models, age increase, intra-amniotic infection of the mother together with IL-6 increase showed to be positively alarmed with longer hospital days (longer than 7 days), while cough sign seemed to have a negative indication, which could be used as a reference in evaluating the prognosis.
Although progress has been made in the reduction of morbidity and mortality from neonatal sepsis, we still lack accurate diagnostic tools for neonatal sepsis, complicating the management of this condition (16). Sepsis in neonates remains one of the most significant causes of morbidity and mortality, especially for preterm newborns in intensive care units (17). The clinical characteristics-oriented predictive model could set rapid care alarms, which could be significant (16, 17). In this study, 2 pre-diagnostic models and 1 prognosis predictive model had excellent performance, which could be suggested as useful pre-diagnostic tools and a novel therapeutic strategy for neonatal sepsis.
From a methodological perspective, overfitting is often observed when there is high variance in the development of predictive models. Overfitting is related to the model complexity or inadequate size of the training data. To avoid overfitting, cross-validation was conducted to assess the model's fit and to determine its accuracy. The AUC curve was also used to assess the model performance, and the results showed excellent sensitivity and specificity.
5.1. Conclusions
Good performance of diagnostic and prognostic models was established for neonatal sepsis reference, together with therapeutic strategy guiding marker. We hope the findings from this study could offer clinical help in the diagnosis and therapy of neonatal sepsis.