1. Background
Group B streptococcus (GBS), also known as Streptococcus agalactiae, is usually found in the female reproductive tract and can cause endometritis and chorioamnionitis in pregnant women via the urinary tract (1, 2). It is also a common bacterial pathogen causing infections in newborns, such as sepsis, meningitis, and pneumonia, following exposure in an infected birth canal (3). Neonatal GBS infections have two major forms, early-onset disease (EOD) that occurs within the first 7 days after birth and late-onset disease (LOD) that occurs later than 7 days after birth (4, 5). As a common bacterial pathogen causing neonatal infections in Western countries, GBS is also a major causative agent of neonatal infections in China, with a subsequent high mortality and morbidity. A number of virulence factors, such as specific capsular polysaccharide (CPS), lipoteichoic acid, hyaluronic acid, and neuraminidase, are involved in GBS pathogenicity (6). A total of 10 GBS serotypes (Ia, Ib, II, III, IV, V, VI, VII, VIII, and IX) have been characterized based on the antigenicity of CPS X (7, 8). Epidemiological studies have demonstrated that strains of serotype III caused 86.2% of meningitis and 60.8% of sepsis in the study group (6). Therefore, CPS-based serotypes may be associated with the pathogenesis of invasive GBS.
Multilocus sequence typing (MLST), a powerful genotyping tool based on a number of housekeeping genes, has been widely used in clinical microbiology and epidemiological studies (9). Genetic polymorphisms of GBS strains are independent of CPS-based serotypes (10). Tazi et al. reported that GBS strains of the same serotype exhibited different genotypes, which were associated with distinct clinical manifestations (11). ST-17 has been shown to be associated with GBS LOD and meningitis (12). In addition, the sequence types ST17 and ST10 are associated with meningitis, and sequence types ST1 and ST23 are associated with septicemia (13, 14).
Since 1970s, GBS infections have been widely investigated in Western countries. Vaginal and anal swabs for GBS detection and characterization are a standard prenatal procedure, thereby producing an accurate picture of GBS occurrence in pregnant women and infants. In China, GBS detection is not a compulsory prenatal test. Therefore, it is important to further the understanding of GBS infection, genotypic distribution, serotypes, and antibiotic resistance in China.
2. Methods
2.1. GBS Isolation and Identification
Between January 2008 and August 2014, a total of 26 GBS strains were isolated from throat swabs, sputum, umbilical secretions, or blood of 26 neonates, including 12 males and 14 females hospitalized in the maternal and child health hospital of futian District, Shenzhen and Shenzhen Municipal people’s hospital. The isolated GBS strains were identified and characterized using the Christie-Atkins-Munch-Petersen (CAMP), automated microbial identification system, and the VITEK2 Compact (BioMerirux, France). The GBS isolates were stored at -80°C for follow-up experiments.
2.2. Serotyping and Evaluation of Antimicrobial Susceptibility
Serotyping of the 26 GBS strains was conducted using the Brochure STREP-B-LATEX latex agglutination serotyping kit (SSI, Denmark) according to the manufacturer’s instructions.
The antimicrobial susceptibility of the 26 strains was evaluated using the disk diffusion method and an E-test. The drug sensitivity test strips for the oxcomycin, tetracycline, chloramphenicol, levofloxacin, minocycline, penicillin, ceftriaxone, cefepime, and Vancomycin tests were purchased from the Oxoid company. The E-test results were interpreted according to the American society for clinical laboratory standards (CLSI) (http://clsi.org/), in order to determine the antimicrobial susceptibility (susceptible, sensitive, and intermediate levels), as well as the minimum inhibitory concentration (MIC).
2.3. Isolation of Genomic DNA and MLST Analysis
Genomic DNA was isolated from all 26 GBS strains using the TianGen Genomic DNA isolation kit according to the manufacturer’s instructions, and was stored at 4°C for MLST analysis.
A total of seven housekeeping genes (adhP, pheS, atr, glnA, sdhA, glcK, and tkt) were included in the MLST analysis. The primers used for the amplification of the seven housekeeping genes are listed in Table 1. PCR amplicons were sequenced with BGI and the sequences were analyzed using Chromas software. The DNA sequences of the seven housekeeping genes were aligned with the agalactiae MLST database to determine the sequence type (ST).
| Gene | Primer | Primer Sequence (5’ - 3’) | Amplicon Size, bp |
|---|---|---|---|
| adhP | Forward | GTTGGTCATGGTGAAGCACT | 704 |
| Reverse | ACTGTACCTCCAGCACGAAC | ||
| pheS | Forward | GATTAAGGAGTAGTGGCACG | 762 |
| Reverse | TTGAGATCGCCCATTGAA | ||
| atr | Forward | CGATTCTCTCAGCTTTGTTA | 778 |
| Reverse | AGAAATCTCTTGTGCGGAT | ||
| glnA | Forward | CCGGCTACAGATGAACAATT | 709 |
| Reverse | CTGATAATTGCATTCCACG | ||
| sdhA | Forward | AGAGCAAGCTAATAGCCAAC | 684 |
| Reverse | ATATCAGCAGCAACAAGTGC | ||
| glcK | Forward | CTCGGAGGAACGACCATT | 641 |
| Reverse | CTTGTAACAGTATCACCGTT | ||
| tkt | Forward | CCAGGCTTTGATTTAGTTGA | 657 |
| Reverse | AATAGCTTGTTGGCTTGAAA |
Primers Used for Amplification of Seven Housekeeping Genes
2.4. Statistical Analyses
Statistical analyses were conducted using the SPSS 13.0 package. Antibiotic resistance was evaluated on the basis of the percentage of strains resistant to a given drug. The chi-square test was used and a P value < 0.05 was considered to be statistically significant.
3. Results
3.1. Basic Characteristics of the Patients
The age of the 26 neonates ranged from 0 to 30 days with an average of 8.615 days. Of the 26 neonates, 23 were term and three premature. Twenty-five children were within the normal weight range, and one had low-birth weight. In addition, eight neonates were born by cesarean section, and 18 had vaginal birth. Premature rupture of membranes was reported in four cases and diabetes mellitus was reported in four of the mothers.
3.2. Four Serotypes Were Identified Among the 26 GBS Strains
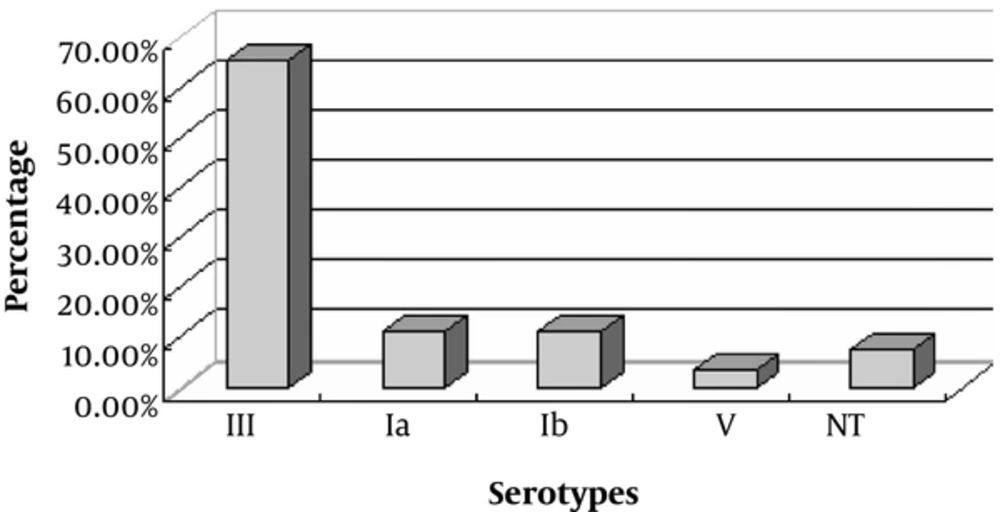
Based on the VITEK identification system, biochemical tests, and CAMP, 26 isolates were identified as GBS strains. Four serotypes, including Ia, Ib, III, and V, were identified from the 26 strains. Of these, 3 (11.5%), 3 (11.5%), 17 (65.4%), and 1 (3.8%) were identified as serotypes Ia, Ib, III, and V, respectively. In addition, 2 (7.7%) GBS strains were untypable (Figure 1).
3.3. Antibiotic Resistance
All 26 GBS isolates were susceptible to minocycline, penicillin, ceftriaxone, cefepime, vancomycin, meropenem, and chloramphenicol. In addition, 84.6% of GBS strains were susceptible to levofloxacin. All isolates were resistant to tetracycline. In addition, 84.6% and 81.8% of GBS strains were resistant to erythromycin and clindamycin, respectively. The MIC of erythromycin was > 256 mg/L and the MIC of clindamycin ranged from 32 to 256 mg/L. The MIC of tetracycline ranged from 12 to 48 mg/L, that of levofloxacin was 0.75 mg/L, that of penicillin ranged from 0.032 to 0.064 mg/L, and the MIC of minocycline ranged from 4 to 8 mg/L (Tables 2 and 3).
| Antibiotics | Susceptible, % | Intermediate Resistant, % | Resistant, % |
|---|---|---|---|
| Tetracycline | 0 | 0 | 100 |
| Erythromycin | 15.40 | 0 | 84.60 |
| Clindamycin | 18.60 | 0 | 81.80 |
| Levofloxacin | 84.60 | 0 | 15.40 |
| Minocycline | 100 | 0 | 0 |
| penicillin | 100 | 0 | 0 |
| Ceftriaxone | 100 | 0 | 0 |
| Cefepime | 100 | 0 | 0 |
| Vancomycin | 100 | 0 | 0 |
| Meropenem | 100 | 0 | 0 |
| Chloramphenicol | 100 | 0 | 0 |
Antimicrobial Susceptibility of the 26 GBS Strains Isolated from Neonates in China
| Antibiotics | MIC50, mg/L | MIC90, mg/L | MIC range, mg/L | Susceptibility, % | Intermediate Resistant | Resistant, % |
|---|---|---|---|---|---|---|
| Erythromycin | > 256 | > 256 | 0.25 - > 256 | 15.40 | 0 | 84.60 |
| Clindamycin | 32 | > 256 | 0.094 - > 256 | 18.20 | 0 | 81.80 |
| Tetracycline | 24 | 48 | 12 - > 256 | 0 | 0 | 100 |
| Levofloxacin | 0.75 | > 32 | 0.5 - > 32 | 84.60 | 0 | 15.40 |
| Penicillin | 0.064 | 0.064 | 0.032 - 0.064 | 100 | 0 | 0.00 |
| Minocycline | 4 | 8 | 0.8 - 8 | 100 | 0 | 0.00 |
The Minimum Inhibitory Concentration (MIC) of 6 Commonly-Used Antibiotics for 26 GBS Isolatesa
3.4. Genotypic Distribution of GBS Strains Based on MLST
A total of 8 sequence types, including ST17, ST12, ST19, ST171, ST456, ST485, and ST23 were identified among the 26 GBS strains. One (3.8%) isolate was untypable based on MLST. ST17 was identified in 13 (50%) isolates. In addition, ST12, ST19, ST171, ST456, and ST23 were identified in 4 (15.4%), 4 (15.4%), 1 (3.8%), 1 (3.8%), and 1 (3.8%) GBS strains, respectively (Table 4).
3.5. The Association Between Serotypes, Genotypes, and Clinical Manifestations of GBS Strains
Of the 26 cases of GBS infection, there were two cases of omphalitis (one isolate of serotype Ib and one isolate of serotype V), six cases of pneumonia (one isolate of serotype Ia, one isolate of serotype Ib, two isolates of serotype III, and two isolates with an untypable serotype), one case of septicemia (serotype III), eight cases of sepsis with pneumonia (serotype III), three cases of sepsis with purulent meningitis (one isolate of serotype Ia and two isolates of serotype III), and six cases of sepsis with pneumonia and purulent meningitis (one isolate of serotype Ia, one isolate of serotype Ib, and four isolates of serotype III). Of the 18 GBS strains associated with sepsis, serotype III was identified in 15 strains (83.3%, P < 0.05). Of the 9 GBS strains associated with purulent meningitis, serotype III was identified in 6 GBS strains (66.7%, P < 0.05). Of the 20 GBS strains associated with pneumonia, serotype III was identified in 14 GBS strains (70%, P < 0.05) (Table 5). Therefore, serotype III GBS strains were the major cause of sepsis, purulent meningitis, and pneumonia through GBS infections in newborns.
| Clinical Manifestation | Sampling Site | Number of Isolate | Serotype | Genotype |
|---|---|---|---|---|
| Omphalitis | Umbilical secretions | 2 | Ib (1), V (1) | ST12 (1), ST456 (1) |
| Pneumonia | Blood, throat swab, sputum | 6 | Ia (1), Ib (1), III (2), NT (2) | ST12 (1), ST19 (1), NT (1), ST17 (2), ST485 (1) |
| Sepsis | Blood | 1 | III (1) | ST17 (1) |
| Sepsis with pneumonia | Blood | 8 | III (8) | ST17 (6), ST171 (1), ST19 (1) |
| Sepsis with purulent meningitis | Blood | 3 | Ia (1), III (2) | ST17 (2), ST23 (1) |
| Sepsis with pneumonia and purulent meningitis | Blood | 6 | Ia (1), Ib (1), III (4) | ST17 (2), ST19 (2), ST12 (2) |
The Association Between Serotypes, Genotypes, and Clinical Manifestations of the GBS Strains
Sequence types ST17, ST12, ST19, ST171, ST456, ST485, and ST23 were identified in 13 (50%), 4 (15.4%), 4 (15.4%), 1 (3.8%), 1 (3.8%), 1 (3.8%), and 1 (3.8%) GBS strains, respectively. In addition, one (3.8%) GBS strain was not typable. The nine cases of purulent meningitis consisted of 4 (44.4%, P < 0.05) ST17, 2 (22.2%) ST12, 2 (22.2%) ST19, and 1 (11.1%) ST23 strains. The 18 cases of sepsis were caused by 11 (61.1%, P < 0.05) ST17, 2 (11.1%) ST12, 3 (16.7%) ST19, and 1 (5.6%) ST12 strains. The 20 cases of pneumonia were caused by 10 (50%, P < 0.05) ST17, 3 (16.7%) ST12, 4 (22.2%) ST19, 1 (5.6%) ST485, and 1 (5.6%) untypable GBS strains (Table 5). Taken together, ST17 was the major genotype causing sepsis, purulent meningitis, and pneumonia through invasive GBS infections in newborns.
4. Discussion
Group B Streptococcus (GBS) is a Gram-positive bacterium of the B group, according to the Lancefild serological classification. GBS strains have become the most common causative agent of neonatal and infant infections in Europe and the United States since 1970s (3). In recent years, the rate of GBS infection has gradually increased in China with the strict control of antibiotics. According to the specificity of capsular polysaccharide antigens, at least 10 serotypes have been identified in GBS strains (15). The infection rate and the prevalence of different GBS serotypes are distinct among different regions, ethinicities, sampling sites, detection methods, and timepoints (16, 17). In general, strains of serotypes III, Ia, Ib, II, and V cause the most GBS infections (18, 19). In the present study, we found that 17 (65.4%), 3 (11.5%), 3 (11.5%), and 1 (3.8%) strains were serotypes III, Ia, IIb, and V, respectively. In addition, two (7.7%) strains were untypable. Therefore, serotype III was the major cause of GBS infection in our hospital, which is consistent with previous studies. Of the 18 cases of septicemia, 15 were caused by serotype III. Of the 9 cases of purulent meningitis, 6 were caused by serotype III. In addition, of the 20 cases of combined pneumonia, 14 were associated with serotype III. These results suggest that GBS serotype III is the most common serotype causing septicemia, purulent meningitis, and combined pneumonia, which is consistent with a previous study (15). In this study, the authors found that GBS strains serotypes III and Ia were the major serotypes causing invasive infections in newborns. An epidemiological study conducted in Europe and the United States found that 86.2% of meningitis and 60.8% of sepsis cases were caused by GBS serotype III strains (6).
In the present study, we explored the genotypic distribution of 26 GBS strains. ST17 was identified in 13 strains (50%), suggesting that ST17 is the major genotype causing invasive GBS infections in newborns. Of the nine cases of purulent meningitis, four (44.4%) were caused by ST17 genotype. Of the 18 cases of septicemia, 11 (61.1%) were caused by ST17 genotype. Of the 20 cases of pneumonia, 10 (50%) were caused by ST17 genotype. Taken together, ST17 is the major genotype associated with septicemia, purulent meningitis, and pneumonia. Following ST17, ST12 and ST19 are the next most common genotypes causing GBS infections in newborns, a finding consistent with those reported by Jones et al. (9). In this study, the authors reported that sequence types ST17, ST19, ST23, and ST1 were the major genotypes causing invasive GBS infections in newborns. According to a study conducted by Alkuwaity et al., in the French national reference center, ST17 is the most common genotype among 651 GBS strains. In addition, ST17 was associated with over 80% of meningitis cases.
For a long period, β-lactam antibiotics have been considered as the first choice of antibiotics for the treatment of GBS infections. For patients allergic to β-lactam antibiotics, macrolide antibiotics are used as alternatives. Gygax et al. reported that resistance to macrolide antibiotics is an increasing problem for the treatment of GBS infections (20). In the present study, we found that all GBS strains were susceptible to penicillin, ceftriaxone, and cefepime, suggesting that penicillin should remain the first-line antibiotic for the treatment of GBS infection. All strains were also susceptible to chloramphenicol, minocycline, vancomycin, and meropenem. In addition, 84.6% of the strains were susceptible to levofloxacin. Finally, 100%, 84.6%, and 81.8% of GBS strains were resistant to tetracycline, erythromycin, and clindamycin, respectively. It has been reported that 50.7% and 38.4% of GBS strains were resistant to erythromycin and clindamycin, respectively, in the United States (21). According to a study conducted by Li et al. in China, 55.21%, 43.7%, and 93.75% of 96 GBS isolates were resistant to erythromycin, clindamycin, and tetracycline, respectively (22). No significant differences in the rates of erythromycin and clindamycin resistance were identified among GBS strains isolated from perinatal infections, GBS early-onset disease, and late-onset disease (23). However, different rates of resistance to macrolide antibiotics were found between distinct countries and regions, suggesting that the resistance of different GBS strains to macrolide antibiotics is associated with ethnicity. Typically, GBS strains have high rates of resistance to tetracycline, erythromycin, and clindamycin, which usually cause a number of adverse reactions; therefore, these are not used as first-line antibiotics for the treatment of GBS infections. For infected patients who are allergic to penicillin and cephalosporin antibiotics, other antibiotics with low resistance rates should be considered. Given that chloramphenicol causes gray baby syndrome, this study suggests antibiotics like vancomycin, minocycline, or meropenem.
Nevertheless, our study has some limitations. Firstly, the sample size was too small, leading to insufficiently rich clinical data. More experiments are needed to validate our conclusions. We will therefore collect more cases and conduct further research in the future.
4.1. Conclusion
In summary, we isolated and characterized 26 GBS strains from newborn infections. Most strains were identified as serotype III and genotype ST17, which was responsible for the majority of sepsis, purulent meningitis, and pneumonia in this study. In addition, penicillin should still be the first-line antibiotic for treating GBS infections. Our study is useful for the diagnosis and treatment of GBS infections in newborns.