1. Background
Neonatal hyperbilirubinemia (NH), a common condition in newborns, occurs in up to 60% of healthy full-term infants (1). This clinical issue arises when the total serum bilirubin (TSB) level exceeds 5 mg/dL, potentially progressing to severe neonatal jaundice (2). In such cases, NH can lead to neurodevelopmental complications, including hearing loss, cognitive impairment, and athetosis. In its most severe form, it may result in seizures, coma, or death.
It has been proposed that genetic, demographic, and environmental factors contribute to the risk of NH (3, 4). Genetic variations, in combination with other icterogenic factors, may significantly increase susceptibility to NH (5, 6). Among these genetic contributors, uridine diphosphate glucuronosyltransferase 1A1 (UGT1A1) has been strongly associated with NH (7, 8). UGT1A1 plays a crucial role in bilirubin conjugation (9). The gene is located on chromosome 2q37, and polymorphisms within its coding region or promoter can lead to reduced enzyme activity. This enzymatic deficiency results in elevated levels of unconjugated serum bilirubin, which manifests clinically as hyperbilirubinemia-related conditions such as Gilbert’s syndrome (GS) and Crigler-Najjar syndrome (CNS) (9-11).
Several polymorphisms of UGT1A1 have been identified in GS and CNS, including Gly71Arg and variants in the TATA-box promoter region (12, 13). In addition to these well-known mutations, other polymorphisms such as rs3755319 A/C and rs201295078 (14-bp insertion/deletion, I/D) also exist within the UGT1A1 gene. Associations between the rs3755319 polymorphism and conditions such as congenital heart disease (14), colorectal cancer (15), and tuberculosis (16) have been explored in previous studies.
While the roles of the UGT1A1 Gly71Arg and TATA promoter polymorphisms in NH have been widely studied across various populations and ethnic groups (1, 17-20), no published case–control study has yet evaluated the association between UGT1A1 rs3755319 and rs201295078 variants and susceptibility to NH.
2. Objectives
In the current case–control study, we aimed to explore the correlation between UGT1A1 rs3755319 and rs201295078 gene polymorphisms and neonatal hyperbilirubinemia (NH) in an Iranian population.
3. Methods
3.1. Subjects
This case–control investigation was conducted on 110 newborns with hyperbilirubinemia (cases) and 112 healthy newborns (controls) at Ali-Ebne-Abitaleb Hospital, affiliated with Zahedan University of Medical Sciences, Zahedan, Iran. The study received ethical approval from the local Ethics Committee of Zahedan University of Medical Sciences (IR.ZAUMS.REC.1391.5795). Prior to participation, written informed consent was obtained from the parents of all subjects. As previously described, genomic DNA was isolated from peripheral blood samples (21).
3.2. Genotyping
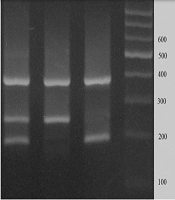
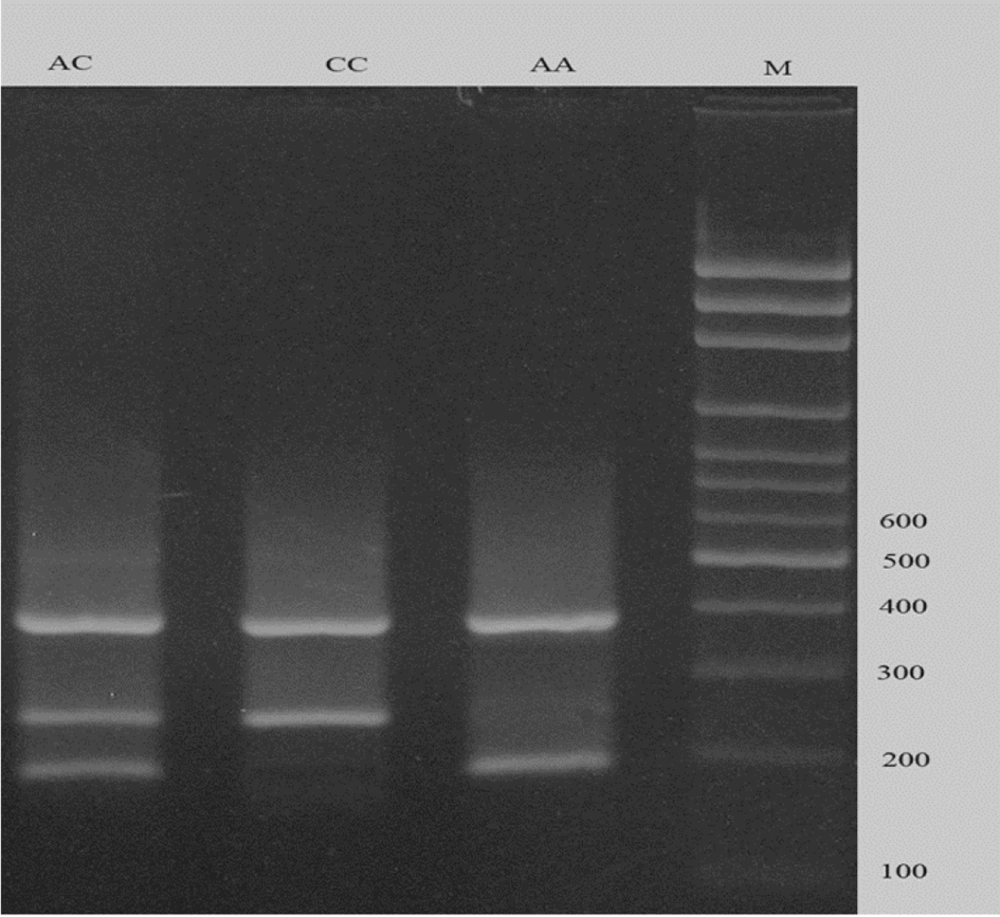
To genotype the rs3755319 A/C polymorphism, the tetra-primer amplification refractory mutation system polymerase chain reaction (T-ARMS-PCR) method was employed. Primer sequences are listed in Table 1. The product sizes were 192 bp for the A allele, 254 bp for the C allele, and 390 bp for the two outer primers (control band) (Figure 1). Each 20 μL PCR reaction contained 1 μL of DNA (approximately 100 ng/μL), 1 μL of each primer, 10 μL of 2X Prime Taq Premix, and 5 μL of double-distilled water (ddH2O were added. The conditions for PCR cycling were determined as follows: Five minutes initial denaturation at 95°C, followed by 30 cycles consisting 30 seconds denaturation at 95°C, 30 seconds annealing at 57°C, and 30 seconds at 72°C for extension. A final extension was established at 72°C for 5 minutes. In order to analyze the PCR results, electrophoresis on a 2% agarose gel supplemented with ethidium bromide was used. Subsequently results were visualized under UV).
| Polymorphism | PCR primers (5'→3') | Method |
|---|---|---|
| UGT1A1 (rs3755319) | T-ARMS-PCR | |
| FO | TGGGATCTGTGCAGTTATCTTGGAATTG | |
| RO | CCTTGTGTTCTGTGGAACTCACCTTCAT | |
| FI (C allele) | TGCTCATCTTTCCCTTTTGACTTCGAC | |
| RI (A allele) | AGGCATTTGGGGAAATTCTGATGACTAAT | |
| UGT1A1 (rs201295078 14-bp I/D) | PCR | |
| F | ATCGTGTGGGTCCTGAATTGG | |
| R | TATTCCCAGTCAGAGGCGCTA |
The Primers Used for Detection of UGT1A1 rs3755319 and rs201295078 Gene Polymorphisms
The PCR cycling conditions were as follows: initial denaturation at 95°C for 5 minutes, followed by 30 cycles of 30 seconds at 95°C (denaturation), 30 seconds at 57°C (annealing), and 30 seconds at 72°C (extension). A final extension step was performed at 72°C for 5 minutes. PCR products were analyzed by electrophoresis on a 2% agarose gel containing ethidium bromide, and bands were visualized under UV illumination.
3.3. PCR Conditions for rs201295078
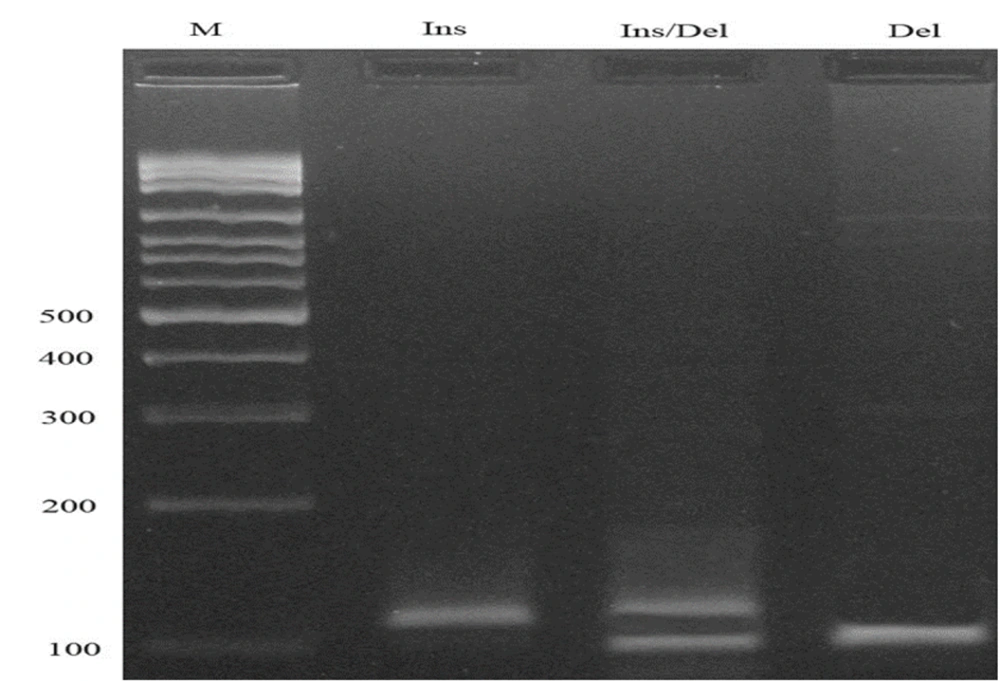
The rs201295078 (14-bp insertion/deletion, I/D) polymorphism was detected using a conventional PCR method. PCR cycling conditions included an initial denaturation at 95°C for 5 minutes, followed by 30 cycles of 30 seconds at 95°C (denaturation), 27 seconds at 66°C (annealing), and 30 seconds at 72°C (extension), with a final extension at 72°C for 10 minutes. The resulting fragment sizes were 114 bp for the insertion allele and 100 bp for the deletion allele (Figure 2).
3.4. Statistical Analysis
All statistical analyses were performed using SPSS version 21.0. The independent samples t-test was used to analyze continuous variables, and the chi-square (χ2) test was employed for categorical data. Logistic regression was used to evaluate the association between UGT1A1 rs3755319 and rs201295078 polymorphisms and the risk of NH by calculating odds ratios (OR) with 95% confidence intervals (CI). A P-value < 0.05 was considered statistically significant.
3.5. In Silico Analysis
TFBIND was utilized to assess the potential impact of single nucleotide polymorphisms (SNPs) on transcriptional regulation. This tool predicts transcription factor binding sites (TFBSs) within a DNA sequence by comparing it to known consensus motifs. The output includes the predicted transcription factor name, binding position, strand orientation, and a similarity score ranging from 0 to 1. Higher similarity scores indicate a stronger match between the input sequence and the TFBS consensus, suggesting a greater likelihood of transcription factor binding. By comparing the scores of wild-type and mutant alleles, one can infer whether a specific SNP enhances or disrupts transcription factor binding, potentially influencing gene expression (22).
4. Results
Totally 222 neonates including 110 neonates (ages: 6.1 ± 2.1days; male/female = 54/56) with and 112 without hyperbilirubinemia (ages: 6.8 ± 2.9 days; male/female = 54/58) enrolled in the study. No significant difference was detected in age and gander across the groups (P = 0.378 and 0.793, respectively). Genotypes and allelic frequencies of the variants are shown in Table 2. The results of this study showed that the UGT1A1 rs3755319 increased the risk of hyperbilirubinemia within codominant (OR = 8.23, 95%CI = 2.31 - 25.18, P < 0.001, AA vs CC), recessive (OR = 4.25, 95% CI = 4.87 - 41.65, P < 0.001, AA vs CC+CA) genetic model. The A allele is associated with increased risk of hyperbilirubinemia (OR = 1.80, 95%CI = 1.23 - 2.62, P = 0.002) in compared to C allele.
| UGT1A1 Variants | Case | Control | OR (95%CI) | P-Value |
|---|---|---|---|---|
| rs3755319 | ||||
| Codominant | ||||
| CC | 15 (13.6) | 13 (11.6) | 1.00 | - |
| CA | 57 (51.8) | 95 (84.8) | 0.52 (0.23 - 1.14) | 0.142 |
| AA | 38 (34.6) | 4 (3.6) | 8.23 (2.31 - 25.18) | < 0.001 |
| Dominant | ||||
| CC | 15 (13.6) | 13 (11.6) | 1.00 | |
| CA+AA | 95 (86.4) | 99 (88.4) | 0.83 (0.38 - 1.84) | 0.689 |
| Recessive | ||||
| CC+CA | 72(65.4) | 108(96.4) | 1.00 | - |
| AA | 38 (34.6) | 4 (3.6) | 4.25 (4.87 - 41.65) | < 0.001 |
| Allele | ||||
| C | 87 (39.5) | 121 (54.0) | 1.00 | - |
| A | 133 (60.5) | 103(46.0) | 1.80 (1.23 - 2.62) | 0.002 |
| rs201295078 | ||||
| Codominant | ||||
| ins/ins | 80 (72.7) | 79 (70.5) | 1.00 | - |
| ins/del | 28 (25.5) | 30 (26.8) | 0.92 (0.51 - 1.68) | 0.790 |
| del/del | 2 (1.8) | 3 (2.7) | 0.66 (0.11 - 4.05) | 0.652 |
| Dominant | ||||
| ins/ins | 80 (72.7) | 79 (70.5) | 1.00 | |
| ins/del+del/del | 30 (27.3) | 33 (29.5) | 0.90 (0.50 - 1.61) | 0.767 |
| Recessive | ||||
| ins/ins+ins/del | 108(98.2) | 109(97.3) | 1.00 | |
| del/del | 2 (1.8) | 3 (2.7) | 0.67(0.11 - 4.11) | 0.666 |
| Allele | ||||
| Ins | 188 (85.5) | 188 (83.9) | 1.00 | - |
| del | 32 (14.5) | 36 (16.1) | 0.89(0.53 - 1.49) | 0.693 |
Genotype and Allele Frequency of UGT1A1 Gene Variants in Newborns with Hyperbilirubinemia and Control Subjects
It was revealed through our analysis that neither the overall chi-square comparison (χ2 = 2.37, P = 0.123) nor the logistic regression analysis within codominant (OR = 0.92, 95%CI = 0.51 - 1.68, P = 0.790; ins/del vs ins/ins; OR = 0.66, 95%CI = 0.11 - 4.05, P = 0.652; del/del vs ins/ins), dominant (OR = 0.90, 95%CI = 0.50 - 1.61, P = 0.767; ins/del+del/del vs ins/ins), recessive (OR = 0.67, 95%CI = 0.11 - 4.11, P = 0.666; del/del vs ins/ins+ ins/del) showed any correlation between UGT1A1 rs201295078 polymorphism and hyperbilirubinemia (Table 2). The results showed that rs201295078 del allele is not associated with hyperbilirubinemia in comparison with insertion allele (OR = 0.89, 95%CI = 0.53 - 1.49, P = 0.693).
The correlation between UGT1A1 genotypes and serum bilirubin levels is shown in Table 3. No statistically significant associations were observed between serum bilirubin concentration and any of the UGT1A1 gene polymorphisms.
| Parameters | rs3755319 | P-Value | rs201295078 | P-Value | ||||
|---|---|---|---|---|---|---|---|---|
| AA | AC | CC | Ins/Ins | Ins/Del | Del/Del | |||
| Total bilirubin | 19.22 ± 5.47 | 19.34 ± 5.82 | 19.23 ± 4.73 | 0.993 | 19.05 ± 5.77 | 19.84 ± 4.89 | 20.75 ± 4.03 | 0.745 |
| Direct Bilirubin | 0.63 ± 0.24 | 0.62 ± 0.22 | 0.61 ± 0.23 | 0.963 | 0.62 ± 0.23 | 0.66 ± 0.22 | 0.55 ± 0.35 | 0.620 |
| Indirect Bilirubin | 18.58 ± 5.37 | 18.72 ± 5.72 | 18.62 ± 4.6 | 0.992 | 18.44 ± 5.66 | 19.19 ± 4.84 | 20.20 ± 3.68 | 0.749 |
Relationship Between Genotypes of UGT1A1 Gene Polymorphisms and Serum Bilirubin Levels
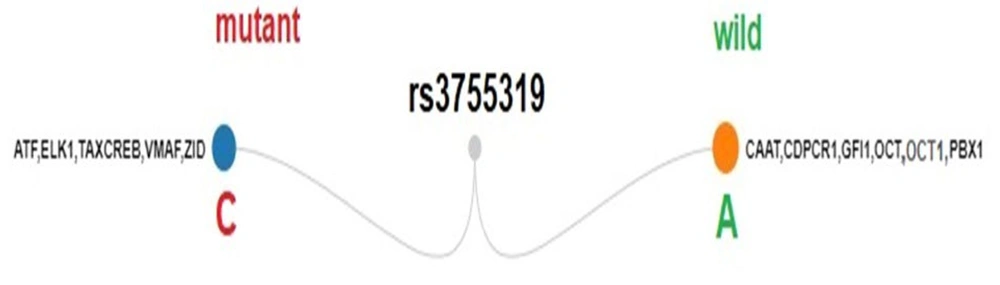
In silico analysis using TFBIND revealed that the rs3755319 SNP may alter transcription factor (TF) binding. A comparison of wild-type and mutant alleles showed that several TFs — including CAAT, CDPCR1, GFI1, OCT, OCT1, and PBX1 — were predicted to bind to the wild-type sequence but lost their binding affinity in the presence of the mutant allele. This loss suggests a potential disruption in transcriptional regulation mediated by these factors. For example, OCT1 is known to modulate gene activation or repression depending on the biological context, while PBX1 is frequently implicated in the regulation of genes involved in cellular differentiation and development (23, 24).
Conversely, TFs such as ATF, ELK1, TAXCREB, VMAF, and ZID, which were not predicted to bind to the wild-type sequence, were identified as putative binding factors in the mutant allele. These newly introduced TF binding motifs may influence gene expression. For instance, ELK1, a transcription factor activated by cellular stress and the MAPK/ERK signaling pathway, typically functions as a transcriptional activator and may contribute to increased gene expression (25, 26) (Figure 3).
5. Discussion
Neonatal hyperbilirubinemia is a common clinical condition affecting newborns (27). The NH results from imbalances in bilirubin synthesis and conjugation, which are primarily due to the immaturity of hepatic metabolic pathways. This leads to increased bilirubin production, impaired hepatic uptake, deficient conjugation, and enhanced enterohepatic circulation. Despite these known factors, the precise etiology of NH remains unclear (28).
Previous research has shown that the UDP-glucuronosyltransferase enzyme plays a crucial role in bilirubin metabolism. This enzyme is encoded by the UGT1A1 gene (29, 30). Genetic polymorphisms in UGT1A1 may occur in various genomic regions, including the promoter, introns, coding exons, splice sites, and distal enhancer elements (31).
Although many studies have evaluated the association between UGT1A1 polymorphisms and NH risk, most of them have focused on two well-known variants: Gly71Arg and the TATA-box promoter polymorphism. Therefore, in the current study, we investigated the correlation between two other UGT1A1 polymorphisms — rs3755319 (A/C) and rs201295078 (14-bp I/D) — and the risk of NH.
Our findings revealed a significant association between the rs3755319 variant and increased susceptibility to hyperbilirubinemia. However, no association was found between rs201295078 and NH. Furthermore, neither polymorphism showed any correlation with serum bilirubin levels.
As previously mentioned, earlier studies have concentrated primarily on the Gly71Arg and TATA-box promoter variants. Some of these investigations found positive associations with NH risk, while others reported no significant findings. In addition to case–control studies, several meta-analyses have been conducted on this topic.
A meta-analysis by Wang et al. included 34 studies — 21 evaluating the Gly71Arg variant and 13 assessing the TATA promoter variant. The results indicated that Gly71Arg was associated with increased NH risk in Asian and African populations, while the TATA promoter variant was linked to NH in Asian and European populations (32).
Similarly, a meta-analysis by Yu et al. identified 32 eligible studies (24 on Gly71Arg and 19 on TATA promoter variants). Their analysis confirmed a significant association between both variants and increased NH susceptibility (33). However, in contrast to these findings, Li and colleagues conducted a meta-analysis that included four studies and found no significant association between the TATA promoter variant and NH risk across allelic, codominant, dominant, or recessive genetic models (34).
Mehrad-Majd et al. (2019) also performed a meta-analysis focusing on the Gly71Arg variant. They included 32 studies and reported that this polymorphism was significantly associated with NH in all genetic models — codominant, dominant, recessive, and allelic. A similar association was confirmed in a subgroup analysis of Asian populations (28).
The rs3755319 polymorphism has also been studied in relation to other diseases. Tao et al. assessed its effect in congenital heart diseases, evaluating four maternal UGT1A1 variants, including rs3755319, and found no significant association (14). Xiao-ling et al. investigated rs3755319 in a study involving 690 colorectal cancer patients and 431 controls, and also reported no significant correlation (15). In contrast, Chen et al. conducted a meta-analysis evaluating UGT polymorphisms and the risk of anti-tuberculosis drug-induced liver injury. Their pooled results demonstrated a significant association between rs3755319 and increased susceptibility to this condition (35).
5.1. Conclusions
Our findings support a significant association between the rs3755319 variant of the UGT1A1 gene and the risk of neonatal hyperbilirubinemia. To validate these results, further studies with larger sample sizes and diverse ethnic groups are recommended. Future research should also explore the role of transcriptional regulatory elements and the MAPK/ERK signaling pathway in the pathogenesis of NH.