1. Introduction
Congenital muscular dystrophy (CMD) is a collection of rare and greatly heterogeneous neuromuscular disorders. The clinical characteristics consist of congenital hypotonia, progressive muscle weakness, muscle contractures, and delayed motor milestones. Merosin deficient congenital muscular dystrophy type 1A (MDC1A) is a kind of CMD with the protein merosin (also known as laminin 2) deficient in the muscle tissue due to mutations in LAMA2 gene located at 6q22 - 23 spanning 65 exons (NM-000426.3). MDC1A holds a predominantly autosomal recessive mode of inheritance. Besides classical features such as muscle weakness, elevated serum creatine kinase (CK), white matter abnormalities on the cranial Magnetic Resonance Imaging (MRI) and specific pattern on muscle biopsy are, also, key characteristics (1).
MDC1A was first described by Tomé FM in 1994 (2). The overall prevalence of CMDs and the percentage of each disorder among CMDs vary markedly in different areas of the world. As for MDC1A, it accounts for about 30% patients with CMD in Europe, 10% in the USA, and only 6% in Japan (3-6). There is no such data in China yet, for it is extremely rare in China with only a few reported cases to date (7, 8). In the present study, we report a case with MDC1A. We have confirmed the diagnosis by clinical presentation, char-acteristic white matter abnormalities, muscle biopsy, and so on as the molecular genetic testing.
2. Case Presentation
The case is a 32-month-old boy from non-consanguineous and healthy parents. In the infant period, he was severely hypotonic and difficult to feed. He raised his head steadily until he was 6 months old and could sit unsupported until he was 24 months old. He could not stand and still cannot. The intellectual and speech devel-opment are normal. He has a 13-month-old sister who can already walk alone without any symptom.
In the physical examination, the boy showed severe axial hypotonia, marked impaired muscle strength, and slight contractures in his elbows and hands.
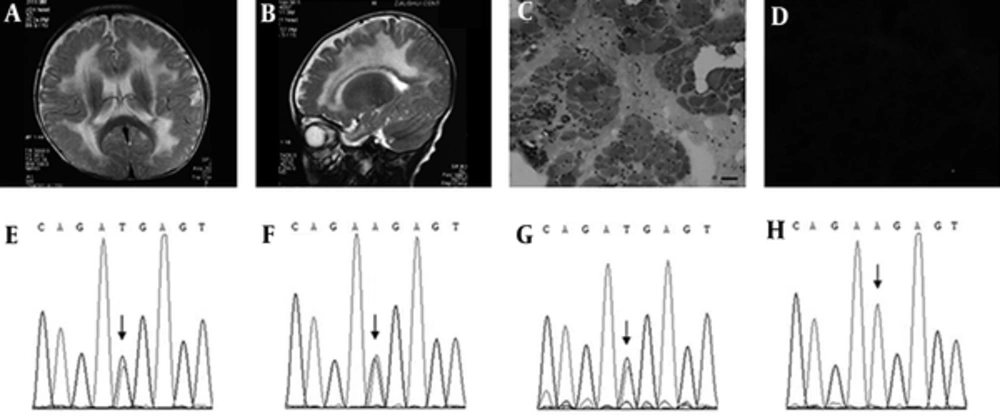
As for blood biochemistry detections, hepatic function, renal function, thyroid gland function, glucose, electrolyte, cholesterol, triglyceride, lipoproteins, ammonia, lactic acid, alkaline phosphatase, and karyotype analysis were all normal. Meanwhile, the CK and CK-MB increased slightly with the results of 370 U/L (normal: 39 - 308 U/L) and 53 U/L (normal: 0 - 25 U/L), respectively. Electroencephalogram was normal for this case. In brain MRI, T1-weighted images showed symmetrical low signal intensity in the white matter of periventricular region, while T2-weighted images revealed remarkable high signal intensity (Figures 1A and B). The manifestation in MRI suggested metabolic brain disease. To exclude this kind of diseases such as leukodystrophy and lysosomal storage disease, we detected the very long chain fatty acid spectrum and the activities of 25 lysosomal enzymes. The results were all normal. To exclude amino acids, fatty acid, and organic acid metabolic diseases, we screened the patient’s blood and urine by tandem mass technology which also showed normal results.
The Results of the Magnetic Resonance Imaging (AB) and the Muscle Biopsy (CD) of the Case and Gene LAMA2 Sequences of the Family (E-H). A, Axial T2-Weighted Image And B, Vertical T2-Weighted Fluid Attenuated Inversion Recovery (FLAIR) Image Show Remarkable Diffuse, Symmetrical High Signal Intensities In The White Matter of Brain, Especially in the Periventricular. HE Staining C, Demonstrates Marked Variations in Fiber Size, Muscle Fiber Necrosis, Irregular Regeneration Arrangement, and Fat Deposits. Immunohistochemical Staining D, With an Antibody to Merosin Reveals That Merosin Is Completely Absent. Bar = 50μm. The Sequencing Exposes A Heterozygous Mutation C.817A>T in the Patient E. Both the Mother F, and the Sister G, Carry This Mutation, While the Father H, Does Not
Needle electromyogram (EMG) showed short spike wave multiphase potential, multiphase irregular waves, and normal nerve conduction velocity, suggesting the damage was myogenic. In biceps brachii muscle biopsy, marked variations in fiber size, prominent connective tissue proliferation, muscle fiber necrosis, regeneration, disordered arrangement, and fat deposit were observed in hematoxylin and eosin staining (Figure 1C). In the immunohistochemical staining with monoclonal antibodies for main proteins in myocyte membrane, all the proteins were detected expressing normally except laminin 2 which was completely absent in the visual fields (Figure 1D). This is just the typical pathological trait of the MDC1A.
To confirm the diagnosis in genetics, genomic DNA of the family was extracted from the blood and LAMA2 genetic analysis was performed by classic Sanger sequencing and multiplex ligation-dependent probe amplification (MLPA). As a result, a maternal point mutation c.817A > T and a paternal exon 4 deletion were discovered for the case. These two variations had never been reported before. The nonsense mutation c.817A > T locates in exon 5, resulting in p.R273X and predicting a truncating laminin 2 protein. Both Mutation Taster and PolyPhen-2 software predicted it to be pathogenic. Both the mother and the sister were heterozygous carriers of this mutation (Figures 1E - 1H). Meanwhile, MLPA analysis verified exon 4 deletion of the LAMA2 gene for the case. The father had the same deletion, while the mother and the sister did not.
3. Discussion
The case displayed typical features of MDC1A. He presented with early hypotonia, weakness, and delayed milestones. The clues for the diagnosis were the elevated CK and typical T2 hyperintensity in the white matter in the brain MRI. The diagnosis was supported by electromyogram and muscle biopsy. Finally, we confirmed the diagnosis by genetic testing.
According to the mode of the autosomal recessive inheritance for this disease, the patient must have held two pathogenic variations in the LAMA2 gene. Sanger sequencing exposed a novel c.817A > T nonsense mutation in exon 5 of the LAMA2 gene from the patient, the mother, and the sister. Potential exonic duplications or deletions were analyzed using MLPA. As a result, the exon 4 deletion was discovered in the LAMA2 gene from the case and his father. Therefore, the asymptomatic sister is a heterozygous carrier sharing the same maternal mutation c.817A > T with the patient, without carrying the paternal exonic deletion.
The manifestation in brain MRI suggested metabolic brain disease. We excluded leukodystrophy, lysosomal storage disease, amino acid metabolic disease, fatty acid metabolic disease, and organic acid metabolic disease by corresponding auxiliary examination. Besides the striated muscle, merosin is also expressed in Schwann cells which are the main cells in nerve myelinization (9-11). Maybe it is why leucodystrophy exists in MDC1A.
A recent review in Lancet gave an updated classification for CMD and figured out the characteristics of MDC1A (12). No clear genotype-phenotype correlation has been established in previous studies. Generally, patients with residual merosin expression often carry deletions in at least one of the two alleles. In contrast, homozygosity or compound heterozygosity for truncating mutations usually result in the absence of merosin and associates with a more severe disease course. And these mutations, more often, localize at the 5’ end of the gene (1). In our case, the point mutation c.817A > T located in exon 5, and the deletion located in exon 4. Both of them were near the 5’ terminal of the LAMA2 gene. Obviously, they would lead to obvious function defects for this protein. The whole merosin protein contained 3210 amino acids, while the broken protein translated by the DNA strain bearing c.817A > T mutation contained 272 amino acids only. Maybe this could explain why the case held such a severe phenotype.
3.1. Conclusion
The diagnosis of MDC1A should be suspected in the presence of an elevated CK, muscle weakness, and abnormal white matter. Diagnosis should be confirmed by muscle biopsy and mutation analysis. The genetic characterization of the affected family will be valuable in prenatal diagnosis and predicting prognosis for siblings. The case we described expands the mutation spectrum of MDC1A.