1. Background
The intestinal microbiota plays an important role in human immunity. The human gut has more than 1,000 species of bacteria living at high densities in the colon (1011 - 1012 cells/g). Currently, many researchers have studied the infants’ microbiota to understand its cellular mechanisms, bacterial composition, and influencing factors but the bacterial colonization is a complex process. In past, until 2006, the healthy neonatal stools were considered sterile at birth (1, 2) except in harmful medical conditions such as amniotic membrane rupture or amnionitis. However, recently the colonization has proved to begin during the prenatal period by identifying the bacteria in amniotic fluid, meconium, and umbilical cord (2, 3). These findings have confirmed that meconium is non-sterile.
The intestine of newborns already contains various bacteria before birth (3). In the beginning, newborns maintain low concentrations of microbiota, but a large shift initiates with introduction of solid foods and becomes similar to those of adults by the age of two (1, 3). Many factors influence this process: delivery method, feeding, gestational age, antibiotics exposure, and prenatal environmental exposure (4). In recent study, a geographical influence has been observed, suggesting “geographical gradient” in the composition of the gut microbiota in Europe (5). Many European studies have shown geographical differences in the composition of microbiota (1, 6-8), but only few studies have been done in East Asia. This study has examined the microbiota composition of healthy newborns born in cities by conducting both culture and molecular analyses. Then, real-time polymerase chain reaction (PCR) and terminal restriction fragment length polymorphism (T-RFLP) analysis have quantitatively detected bacterial groups by their phylogenies (9, 10).
2. Objectives
The aim of this study was to understand and characterize the microbiota composition of healthy Korean newborns in cities.
3. Patients and Methods
128 healthy full-term neonates of both sexes, were born in January 2009 to February 2010 in Seoul St. Mary and St. Paul hospital in Seoul, South Korea. Inclusion criteria were as follows: uncomplicated pregnancy, no congenital abnormality, no pre-and postnatal maternal use of antibiotics, no signs of infection, and no antibiotics treatment of the neonate. We excluded prematurity, low birthweight neonates, and ones who received postnatal antibiotics or any medical treatment. The method of delivery or feeding was not included in this study.
Totally 143 stools were obtained. Second samples were collected from 15 neonates when the first was insufficient or defecated immediately after the first one. Nurses recorded the defecation time of 107 stools. We used 143 samples for bacterial composition and 107 time-recorded samples for bacterial detection by time difference. The stools were put in a stool container and frozen at -80˚C, then transported in an icebox. The Korea Yakult Co. performed fecal culture and molecular ecological analysis. This study follows an approved protocol that follows the guidelines of our institutional review board (IRB) and the amended Declaration of Helsinki.
Samples were put in six culture plates to determine the amounts of total bacteria, anaerobes, gram-positive bacteria, coliforms, lactobacilli, and bifidobacteria, total bacterial count (BHI, Difco), gram-positive bacteria (Columbia, Difco), coliforms (VRB, Difco), anaerobes (WC, Difco), Lactobacillus (MRS with 0.02 % of NaN3, Tokyo Chemical Industry, Japan), and Bifidobacterium (TOS-propionate agar, Yakult, Japan).
In T-RFLP analysis, only 30 samples were used for bacterial DNA extracts because DNA extraction could be performed only in stools with greater than 106 CFU/g. Stools of 50 mg were washed three times with TE buffer (pH 8.0) and the dispersion with 500 µL. It was centrifuged (Hanil, Korea) at 14,000 rpm. After discarding the supernatant, it was washed three times. The mixture of 200 mg of glass beads, 600 µL of TE buffer, 600 µL of phenol, and 100 µL of 10% SDS homogenized twice in Fast-Prep (MP Bio, USA) and was hold at 70 ˚C for 10 min. After 5 min of centrifugation at 4,000 rpm, samples were put with 6 µL of 3 M Na-acetate and 600 µL of isopropanol, then mixed by several inversions. After centrifugation at 14,000 rpm, it was washed with 50 µL of 70% ethanol. The repeatedly with 70% ethanol washed sample was dried at room temperature and added 200 µL of TE buffer.
For purification of template DNA and amplification of 16S rDNA, template DNA was extracted and purified by the manufacturer’s protocol for high pure PCR template preparation kit (Roche, Germany). 16S rDNA region of purified DNA was amplified using universal primer with FAM fluorescent markers, 7F (5’AgAgTTTgATCCTggCTCAg3’) and 1492R (5’ggTTACCTTgTTACgACTT3’) were labeled at 5’. The 50 ng/µL mold of DNA was mixed with each primer. PCR (AccuPower HF PCR Premix, Bioneer, Korea) was initiated 50 µL at 95˚C for 5 minute followed by 30 cycles at 95˚C for 30 second, 50˚C for 30 second, and 72˚C for 90 second for amplification. After 30 times of amplification, the reaction was completed at 72˚C for 10 minutes. Electrophoresis of the amplified PCR was performed with 1% agarose gel at 100 V for 30 minute. Wizard SV Gel and PCR Clean-Up System (Promega) purified the amplified 16S rRNA.
Restriction enzymes, HhaI (TaKara, 2,000 U) and MspI (TaKara, 3,000 U), were used to cut the purified PCR product. HhaI (2.0 µL) was mixed and incubated with 10X M-buffer (1.0 µL). HhaI enzyme (1.0 µL) and deionized water (6.0 µL) were put at 37˚C for 3 hours. MspI (2.0 µL) was mixed and incubated with 10X T-buffer (1.0 µL), MspI enzyme (1.0 µL), BSA (1.0 µL of 0.1%), and deionized water (5.0 µL) at 37˚C for 3 hours.
Data were analyzed using computer software in the Statistical Package for the Social Sciences (SPSS standard version 19.0; IBM SPSS Inc., New York, USA). P values lower than 0.05 were considered statistically significant. In order to find the relationship between Lactobacillus and coliforms, the Spearman correlation coefficient was used. For cluster analysis, BioNumerics ver. 5.0 software (applied Maths, Saint-Martens-Latern, Belgium) interpreted the T-RFLP pattern of the restriction enzymes, HhaI and MspI. The Jaccard similarity and Pearson’s correlation coefficient were used to compare it.
4. Results
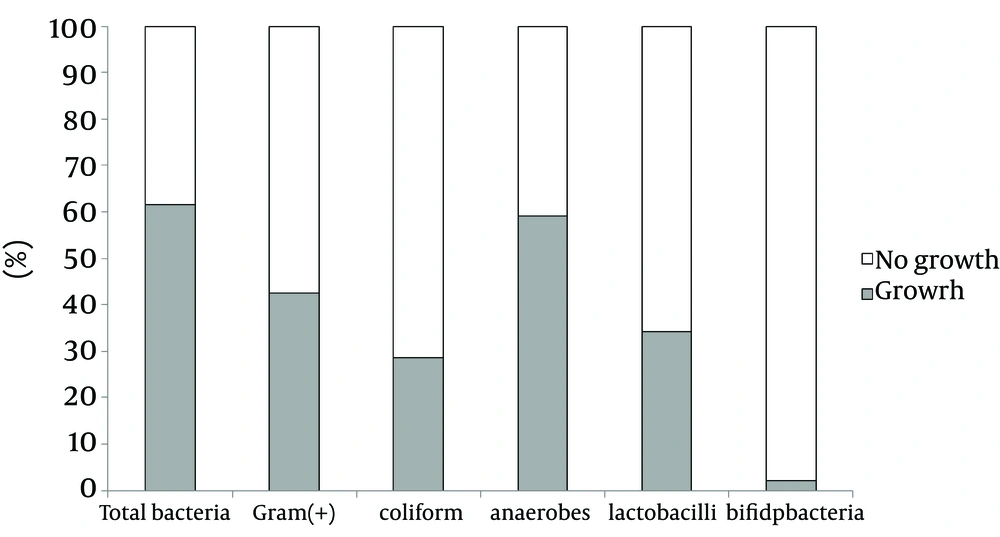
All samples were cultivated in six plates and growth expressed in CFU/g. Total bacteria, anaerobes, gram-positive bacteria, coliforms, lactobacilli, and Bifidobacterium were examined (Table 1). Each stool culture resulted in bacterial counts of about 102 - 109 CFU/g. The total bacterial culture showed 61.5% growth (88/143, range 3.33 × 102 - 3.81 × 109 CFU/g) and 38.5% no growth (Figure 1). Bacterial growth showed anaerobes (59.1%), G + bacteria (42.4%), lactobacilli (34.1%), coliforms (28.5%), and bifidobacteria (2.1%).
| Bacteria | Media | Range (CFU/g) | Mean (CFU/g) |
|---|---|---|---|
| BHI | 3.33 × 102 - 3.81×109 | 1.18 × 108 | |
| WC | 5 × 102 - 3.39 × 109 | 1.21 × 108 | |
| Columbia | 3.2 × 102 - 3.99 × 109 | 7.51 × 107 | |
| MRS | 1.0 × 103 - 1.25 ×109 | 3.77 ×107 | |
| VRB | 3 × 102 - 2.08 × 108 | 1.22 × 106 | |
| TOS | 9 × 103 - 2.3 × 104 | 2.24 × 102 |
a Abbreviations: BHI, brain heart infusion; MRS, Lactobacillus agar acc. to De Man, Rogosa and Sharpe; TOS, propionate agar; VRB, violet red bile; WC, wilkins-chalgren.
b Percentage of bacterial growth from each plate: total bacteria, 61.6%; anaerobes, 59.1%; G (+) bacteria, 42.4 %; lactobacilli, 34.1%; coliform, 28.5%; bifidobacteria, 2.1%.
Within 24 hours after birth, 66.4% (71/107) of the stools passed, whereas 33.6% (36/107) of stools passed after > 24 hours of birth. Before 24 hours, Gram (+) growth was 46.5% (33/71) shown in gray box; no growth in white box, 53.5% (38/71). Coliform growth was 77.8% (28/36) in gray box with no growth of 22.2% (7/36) in white box. There are more bacteria grown in stools passed > 24 hours of birth (P = 0.009).
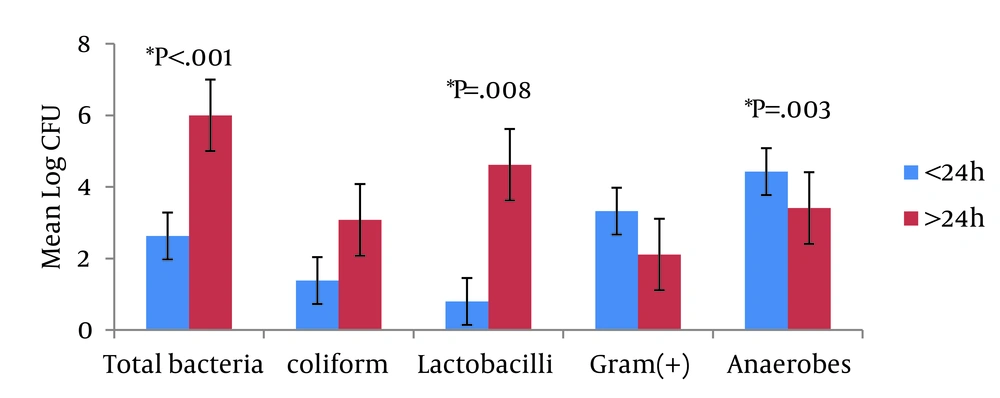
Defecation time was recorded in 107 samples with the latest time being 36 hours and average time 14 ± 7.03 hours after birth. 71 stools (66.4%, range 3.33 × 102 - 1.54 × 108 CFU, average 2.63 ± 2.81 CFU) were defecated within 24 hours after birth. 36 stools (36.6%, range 5.96 × 102 - 3.81 × 109 CFU, average 6.0 ± 3.39 CFU) passed after 24 hours of birth. Significant difference between the growth of bacteria according to the stool defecation time showed in total bacteria (P < 0.001), anaerobes (P = 0.003), and lactobacilli (P = 0.008). Bifidobacteria grew only in two plates (Figure 2). Both coliforms and lactobacilli showed a higher growth in stools > 24 hours after birth (average 3.08 ± 2.85 CFU for coliform, 4.62 ± 3.49 CFU for lactobacilli) than < 24 hours.
The x-axis indicates each culture plate. The y-axis indicates an average count of bacteria in log CFU. The blue bar represents stools passed within 24 hours after birth and red bar represents the stools passed after 24 hours. Total bacteria, lactobacilli, and anaerobes showed statistically significant P values. The vertical bar indicates standard errors.
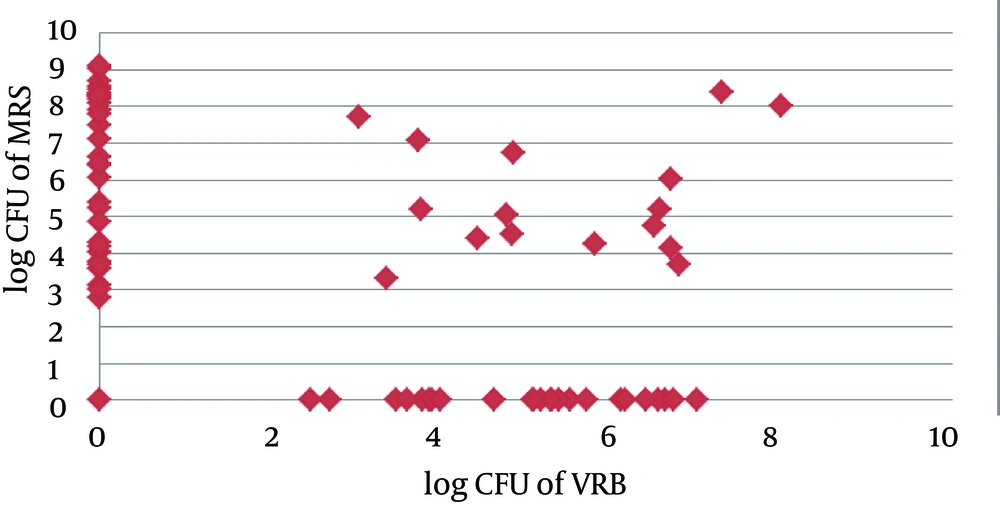
A negative growth correlation between lactobacilli and coliforms has been found. Coliforms were likely to be Gram + bacteria (average 3.32 ± 3.5 CFU) and anaerobes (4.43 ± 3.49 CFU) seemed to be the dominant microbiota in stools passed within 24 hours, although not significant. Coliforms were likely to be found in lactobacilli negative medium. Among 72 samples, 40 stools showed the growth of lactobacilli and 48 stools of coliforms, both growth in 16 stools. The Spearman correlation coefficient rho between the two groups was r = -0.463 (P < 0.001), statistically significant (Figure 3).
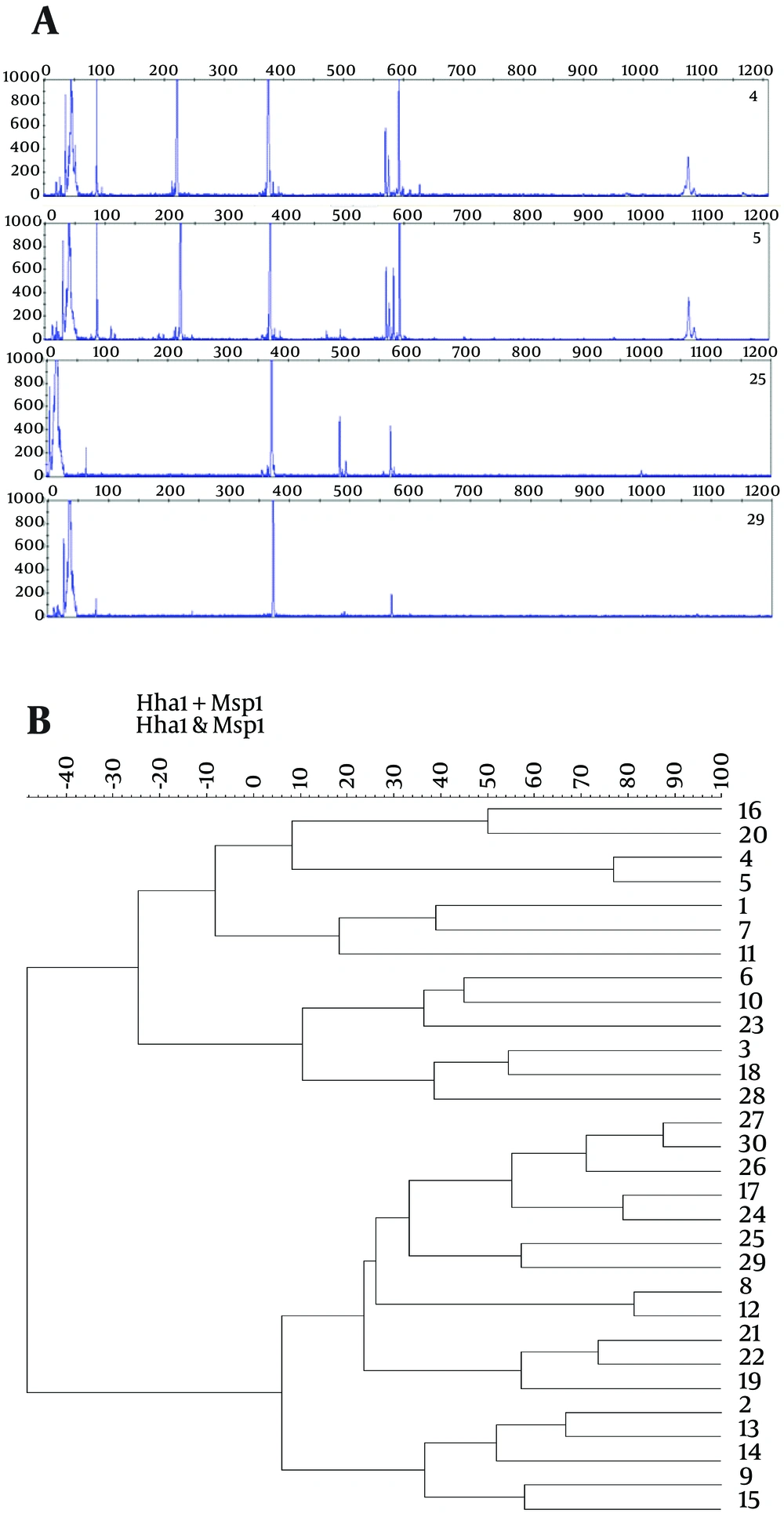
Bacteria diversity was determined by means of T-RFLP extracting bacterial DNA of 30 samples. Each number of peaks represented a measure of diversity. Two restriction enzymes, HhaI and MspI, have been used to identify diversity. Cutting with HhaI, all samples showed peaks around 373 - 375 bp, one showed multiple peaks at 86, 221, and 592 bp with a few other minor peaks. Different patterns of peaks allowed us to differentiate stool samples by their diversity (Figure 4 A). In MspI, all samples showed a peak at 495 bp, many in 70, 79, 565 bp peaks (not shown in data). Numerous peaks showed that the stools have a higher bacterial diversity.
In cluster analysis, the dendrogram was constructed using Ward’s algorithm with 30 samples clustered into two well-defined groups using a hierarchical cluster of HhaI and MspI digestion based on T-RFLP (Figure 4 B). Different bacteria showed different sequences. The upper cluster contained the samples number 16, 20, 4, 5, 1, 7, 11, 6, 10, 23, 3, 18, and 23, while the lower cluster contained the others.
A, T-RFLP patterns of 16S rDNAs from stools digested with HhaI. The 16S rDNAs were extracted from samples and amplified with the universal primers 27F and 1492R. Each peak represents a terminal restriction fragment of a specific length that corresponds to a bacterial phylotype. The meconium sample numbers were 4, 5, 25, and 29. Out of 30 samples, the T-RFLP patterns of 26 samples were similar. B, Hierarchical clustering analysis of microbiota of meconium based on T-RFLP pattern derived from HhaI and MspI digestion. The figure shows the phylogenetic relationships among 30 meconium samples. The x-axis shows the % of similarity and 1 - 30, indicates the sample number.
5. Discussion
The gut of healthy newborns was non-sterile, had inter-individual bacterial profiles with diversity and similarity to infants in western countries. It is the first large molecular study of neonatal microbiota to date in South Korea. To obtain accurate results, we used the most current and accurate microbiological methods to meticulously determine microbiota. Amplification of 16S rRNA is a good method to profile the microbial diversity (11, 12). As expected, most stools (61.5% of total bacterial cultures) were not sterile. In a past Korean study in 2006, the first stools of fifteen healthy Korean newborns were found to be almost sterile (1). Our findings have shown different results. 24 hours after birth, there was a bacterial shift with negative correlation between coliforms and lactobacilli.
By analyzing six culture plates, we found that gram-positive bacteria and anaerobes were dominant flora and early colonizers of the first stool, and this illustrated a typical pattern for gut microbiota (13, 14), while Lactobacillus and Bifidobacterium were less dominant (Table 1). The beneficial microbiota (Lactobacillus and Bifidobacterium) were present in low numbers in the early life of the healthy Korean neonates. A range of facultative anaerobes, such as Enterobacteriaceae, Streptococcus, and Staphylococcus, initially dominated the gut. As the available oxygen is consumed, strict anaerobes, including Bifidobacterium, Bacteroides, and Clostridium proliferate (8). Facultative anaerobes are the first colonizers, but strict anaerobes become abundant later, in which bifidobacteria dominate within days or weeks after birth, ultimately reaching levels that are similar to those of adults by the age of two (15). This result of culture-based study seems to follow the colonization pattern classically described.
Bifidobacterium and Lactobacillus are widely known to be beneficial to intestinal immunity and considered as members of healthy microbiota (16). In general, bifidobacteria colonies by one month of age (7), and the first anaerobes are able to reach high levels in most neonates within the first to second week of life, followed by members of the Firmicutes (17). Bifidobacterium genus has appeared differently in European studies, showing to have geographical influence, more dominant in northern European countries than in southern regions and remained at low colonization rates until day 90 in healthy Greece infants (5, 18). As in other western countries, bifidobacteria in this study have shown a low composition (Table 1).
In different defecation times (Figure 1), greater numbers of bacteria were found in delayed stools. Anaerobes and gram-positive bacteria decreased, while the total number of bacteria increased with the rise of coliforms and lactobacilli after 24 hours. We observed a fast change of composition. In 2009, Savino et al. (3) found that the colonization patterns of gas-forming coliforms, especially E. coli, were more prevalent in colicky infants when compared to healthy infants. In a study comparing the healthy microbiota to allergic colitis, the main differences were in counts of bifidobacteria, clostridia, and total anaerobes with no significant differences in the growth of lactobacilli, coliforms, and enterococci (19). In this study, the increase of Lactobacillus was significant, while the increase of coliforms was insignificant in healthy infants. This result suggests that the dynamics of microbiota could occur in the short time of 24 hours after birth.
We found inhibitory relationship between two bacteria. The negative correlation between lactobacilli and coliform was statistically significant (Figure 3). Coliforms are detrimental to the human gut, have a strong correlation with inflammatory bowel disease and exist highly in infants with necrotizing enterocolitis, Hirshsprung’s disease, and eczema (7, 9, 20). In contrast, lactobacilli are beneficial to gut by promoting adherence of intestinal epithelial cells and inducing mucin secretion in vivo (21). This mechanism inhibits the adherence and invasion of enteropathogenic E. coli and modulates the activation of dendritic cells (22). In plates with Lactobacillus, coliforms were not present or grew in scarce colonies. This could explain the consequence of the earlier colonization of lactobacilli that may play a role in affecting the intestinal immunity to disfavor coliform growth. This suggests that the specific early colonizing bacteria can determine the establishment of healthy gut immunity by their dynamic relationships with other bacteria.
Many studies have already proven that low diversity of gut microbiota at an early age is strongly correlated with the development of inflammatory and allergic disease in an affluent society (23, 24). In the cluster analysis, our findings showed bacterial diversity with different clustering patterns; Enterococcus faecium, Enterococcus durans, Enterococcus hirae, and Streptococcus mutans dominated cluster while the other group did not (Figure 4 A). In a diverse stool, Enterococcus is the dominant flora. This early establishment of diversity explains the existence of inter-individuality in healthy first stools.
The present study has limitation in showing the relation of gut microbiota to onset of disease and lacks clinical information of subjects and other maternal factors. However, many recent studies have found that composition of gut microbiota during early infancy is associated with asthma (25) or inflammatory disease in children. This study confirmed that the healthy microbiota of Korean neonates is non-sterile, in which anaerobes dominate, with low bifidobacteria, and diverse with inter-individuality. Understanding the healthy gut microbiota in infancy, may make it possible to determine core bacteria for ‘healthy microbiota’ throughout life and find strategies to treat conditions associated with a disrupted gut (14). Although the exact mechanism of microbiota in maturation of immunity is elucidated, the “hygiene hypothesis” and the “microflora hypothesis” support that intestinal diversity and different bacterial composition affect the development of inflammatory disease by breaking the balance between Th1 and Th2 (26). The modified “microflora hypothesis” proposes that the overly hygienic and western lifestyle limit general microbial exposure and alter the colonization of the infants’ gut. Overall, this study supports “microflora hypothesis” by showing westernized and various microbiota compositions of the first stools.
This study contributes to further understanding of healthy microbiota and presents preliminary findings to support the “microflora hypothesis” by showing gut composition of city regions in South Korea. The mutually inhibitory mechanism between lactobacilli and coliforms needs further cellular research. Whether and how these different microbiota compositions relate to onset of disease in early infancy need to be further investigated. The development of a more accurate molecular method can strengthen microbiota study. This study paves the way for future study about modulating microbiota of infant into healthy gut.