1. Background
Cystic fibrosis (CF), a life-limiting autosomal recessive disorder, is considered a monogenic disease that is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene (1, 2). However, recent studies have revealed that some patients with a milder CF phenotype do not carry any CFTR gene mutation (3). Located on chromosome 7q31.2, the CFTR gene contains 27 exons and encodes a 1480-aminoacid protein that acts as a cyclic adenosine monophosphate (cAMP)-regulated chloride channel in the apical membrane of epithelial cells (4). Following the identification of the CFTR gene in 1989, it has been found that the ΔF508 mutation is responsible for approximately 70% of the CF cases worldwide (5). To date, more than 1932 mutations have been identified (6). The clinical course of CF includes chronic airway inflammation and recurrent infections that result in progressively deteriorating lung function. The manifestation of CF also includes gastrointestinal problems, growth failure, and male infertility (7). The widely varying phenotypic expression of CF is likely caused by the CFTR allelic heterogeneity and environmental factors (8). The disease variability among patients who share a particular CFTR genotype and similar environment maybe because of additional genetic variations contribute to such variability in the phenotypic expression of CF (9, 10). Genes that interact with the disease-causing mutation are responsible for specific phenotypic alterations and are known as genetic modifiers. Genetic modifiers alter the penetrance, expressivity, pleiotropy, and severity of a phenotype and may play protective roles or increase the susceptibility to disease (11). The gene encoding tumor necrosis factor-α (TNF-α), an endogenous pro-inflammatory cytokine, acts as a modifier gene (8, 9). This gene is located on chromosome 6p21.3, in the class III region of the HLA (12). It has been reported that single nucleotide polymorphisms (SNPs) in the TNF-α promoter region can lead to increased TNF-α production (13, 14). How these polymorphisms alter the susceptibility to systemic lupus erythematosus, insulin-dependent diabetes, and inflammatory bowel disease has been examined (15, 16). Previous studies revealed an association between the number of polymorphic sites of this gene and severe CF phenotype. TNF-α up regulates the activity of other pro-inflammatory cytokines, reduces the concentrations of Growth Hormone (GH), and inhibits the production of insulin-like growth factor 1 (IGF-1) in the skeletal muscle (17, 18). High levels of TNF-α is associated with increased protein catabolism, muscle wasting, and decreased growth and regeneration of skeletal muscle (19). Additionally, TNF-α promotes the neutrophil-dominated inflammatory response, and is inversely correlated with lung function in CF patients (8, 9). The pulmonary phenotype in CF patients is variable including in those who share the same CFTR genotype, and are influenced by secondary genetic factors such as TNF-α gene (20). Regarding the single nucleotide polymorphisms (SNPs) in the promoter region of the TNF-α gene, it has been shown that the -1031C and -308A alleles contribute to increased TNF-α production (13, 14).
2. Objectives
Therefore, the objective of the current study was to investigate the possible relationship between the TNF-α gene 1031T/C and TNF-α 308G/A polymorphisms and the susceptibility to CF disease in patients with no CFTR mutation, and with clinical manifestations in CF patients with homozygous ΔF508 mutation.
3. Materials and Methods
3.1. Subjects
Eighty unrelated Azeri Turkish CF patients were enrolled in this study. Of these patients, 30 were homozygous for ΔF508 and 50 carried no known mutation, but presented the symptoms of CF (samples had been screened for > 90% of all familiar mutations). The diagnostic criteria included positive sweat tests and typical clinical findings of pulmonary and gastrointestinal disease (7, 20). To eliminate the diagnostic variability that might affect the phenotypic categorization, we included only those patients with medical report of clinical symptoms who were referred to our clinic from three hospitals that were located in three separate provinces of north-west Iran. We also studied 157 unrelated, sex-matched, healthy Azeri Turkish control subjects without CF or other inflammatory diseases. The control subjects were not related to any of the patients. The phenotypic characteristics of the patients included gastro intestinal involvement, pulmonary involvement, and poor growth. Meconium ileus complication, rectal prolapse, fat in the stools, bulky and greasy stools, and stomach pain were considered as symptoms related to severe gastrointestinal involvement. Serial infections, excessive inflammation, and bronchiectasis were considered as indicators of more advanced lung disease. Inadequate exocrine pancreas function, weight loss, and malnutrition were regarded as indicators of poor growth (7, 21). The patient classifications were done by specialists.
3.2. DNA Extraction
After receiving approval from the institutional review board and ethical committee of the Tabriz University of Medical Sciences (Tabriz, Iran), approximately 4 mL of intravenous blood samples were collected in EDTA vacutainers and the DNA was extracted using standard methods. All subjects or their parents provided written informed consent for their participation (including the analysis of their DNA) in this study. The samples were deposited in the DNA Bank of the Liver and Gastrointestinal Disease Research Center of Tabriz University of Medical Sciences as anonymous samples.
3.3. Cytokine Gene Polymorphisms
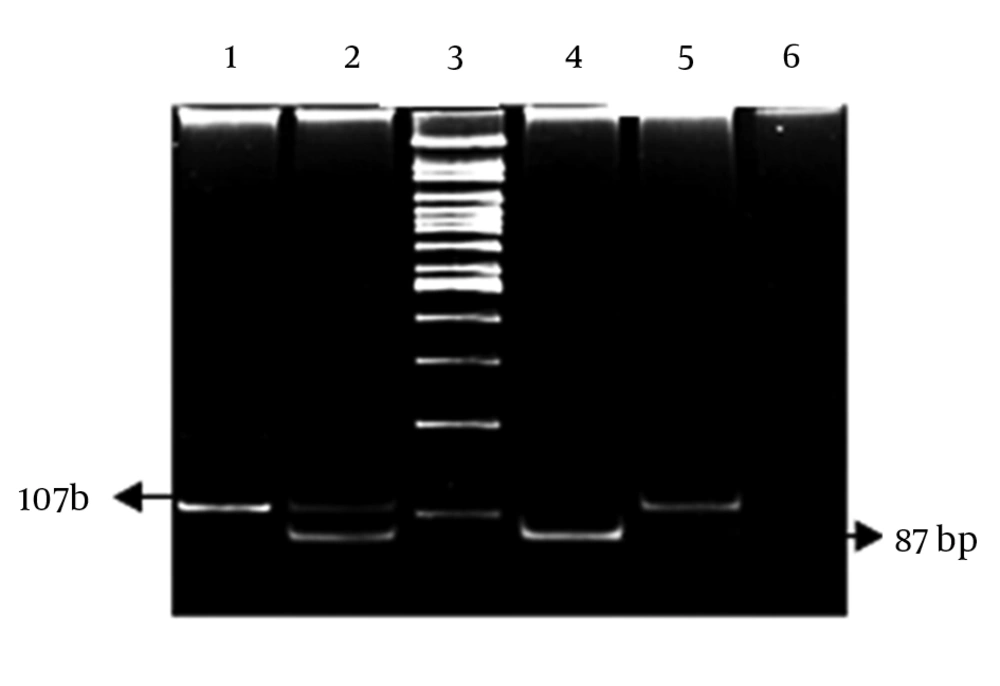
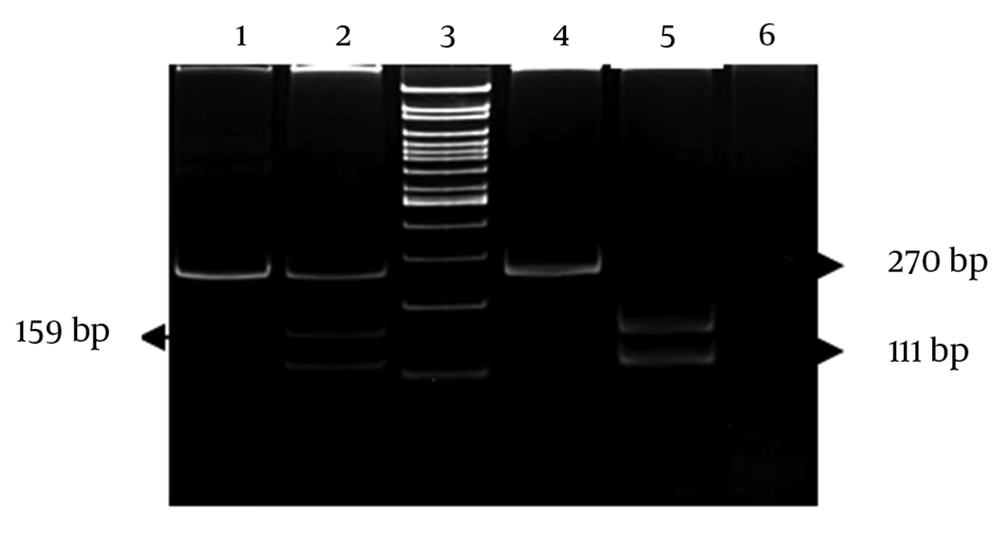
Two fragments of the TNF-α gene promoter, including the -1031T/C and -308A/G polymorphism sites, were amplified by polymerase chain reaction (PCR) (22). The PCR products were digested by incubating with an appropriate restriction enzyme overnight according the manufacturer’s instructions. The digested products were separated on 8% polyacrylamid gels, and the bands were visualized after ethidium bromide staining (Figures 1 and 2).
3.4. Statistical Analysis
The allele, genotype, and haplotype frequencies were compared with the help of the Chi-square test with Yates’ correction or the Fisher’s exact test, wherever appropriate. P < 0.05 was regarded as statistically significant.
4. Results
Data for the 30 patients who were homozygous for CF ΔF508 and classified according to phenotype is presented in Table 1. The frequency of TNF-α gene polymorphic variants were similar in the two groups of patients who did or did not show severe involvement of specific organs and poor growth (Tables 2 and 3). The genotype frequencies of CF patients and healthy controls conformed to the Hardy-Weinberg equilibrium (P = 0.95 for CF patients and P = 0.95 for healthy control subjects). Comparison of allele and genotype frequencies of TNF-α -308G/A and -1031C/T polymorphism showed no significant difference between the patients with non-classic CF symptoms and healthy controls. The frequencies of the TNF-α -308 G/G, G/A, and A/A genotypes were 0.9, 0.1, and 0, respectively in CF patients, and 0.815, 0.165, 0.019, respectively in healthy controls (P = 0.4117 by Fisher’s exact test). TNF-α -308G allele frequencies were 0.95 in CF patients and 0.89 in healthy controls (P = 0.1131, OR = 0.46; 95% CI (0.17-1.22). The allele and genotype distributions of TNF-α 1031T/C were similar in the patient and control groups. The frequencies of the TNF-α 1031 T/T, T/C, and C/C genotypes were 0.66, 0.32, and 0.02, respectively in CF patients, and 0.669, 0.31, 0.19, respectively in controls (P = 0.99 by 2 tests), and the allele frequencies of TNF-α -1031T were 0.82 in CF patients and 0.828 in controls (P = 0.9203, OR = 0.57; 95% CI ( 1.337-1.85). Among the four possible haplotypes, the TNF-α -1031T, -308G was the most frequent in the Iranian Azeri Turkish population. The percentage of the haplotype that predisposes one for having high level of TNF-α (TNF-α -1031C, -308A) was similar in non-classic CF subjects and controls (1.25 vs. 0.95%).
| Total Enrollment | 30 (ΔF508/ΔF508) | Phonotype | Malabsorption | Poor Growth | Pulmonary Involvement |
|---|---|---|---|---|---|
| Severe | 8 (27) | 16 (53) | 15 (50) | ||
| 3 < and 12 > | 6 (20) | ||||
| 12-24 | 9 (30) | ||||
| 24 < | 15 (50) | ||||
| Mild | 22 (73) | 14 (47) | 15 (50) | ||
| Male | 14 (47) | ||||
| Female | 16 (53) | ||||
| 30 (100) |
Demographic and Clinical Characteristics of the CF Patients a
| SNP | Allele Pulmonary Involvement | P Value | Allele Gastro Intestinal Involvement | P Value | Allele Poor Growth | P Value | |||
|---|---|---|---|---|---|---|---|---|---|
| G | A | 0.99 | G | A | 1 | G | A | 1.0 | |
| 29 (96.6) | 1 (3.4) | 30 (93.7) | 2 (6.3) | 15 (93.7) | 1 (6.25) | ||||
| 28 (93.3) | 2 (6.7) | 27 (96.4) | 1 (3.6) | 42 (95.4) | 2 (4.6) | ||||
| T | C | 1 | T | C | 0.12 | T | C | 0.73 | |
| 26 (76.5) | 8 (23.5) | 22 (68.7) | 10 (31.5) | 13 (81.2) | 3 (18.8) | ||||
| 20 (76.9) | 6 (23.1) | 24 (85.7) | 4 (14.3) | 33 (75) | 11 (25) | ||||
| Genotype | Gastro intestinal | Pulmonary | Poor growth | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Severe | Mild | P Value | Severe | Mild | P Value | Severe | Mild | P Value | |
| 1 | 1 | 1 | |||||||
| G/G | 14 (87.5) | 13 (92.8) | 14 (93.3) | 13 (86.6) | 7 (87.5) | 20 (90.1) | |||
| A/G | 2 (12.5) | 1 (8.2) | 1 (6.7) | 2 (13.4) | 1 (12.5) | 2 (9.9) | |||
| A/A | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |||
| 0.5 | 0.7 | 0.47 | |||||||
| T/T | 7 (38.8) | 8 (57.1) | 7 (87.5) | 9 (60) | 5 (62.5) | 11 (50) | |||
| T/C | 9 (61.2) | 6 (42.9) | 8 (12.5) | 6 (30) | 3 (37.5) | 11 (50) | |||
| C/C | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |||
TNF-α Genotype Frequency and Clinical Features of Patients with ΔF508 Mutation in Homozygous State a,b
5. Discussion
To the best of our knowledge, this is the first study of TNF-α gene promoter polymorphisms related to the clinical manifestation of CF in north-west Iran. CF is a pleiotropic disorder with highly variable symptoms. The classic form of CF involves the respiratory tract, gastrointestinal tract, male reproductive tract, and sweat glands and is caused by loss-of-function mutations in the CFTR gene. The non-classic types with milder phenotype are caused by mutations that suppress but do not eliminate the function of the CFTR protein (23). Studies have shown that multiple genes, including TNF-α, act as modifiers in CF. High levels of TNF-α and a disrupted balance between its protective and deleterious roles is frequently observed in CF (24, 25). Previous studies reported that CFTR gene is regulated by cytokines (TNF-α and IL-1) in a cell type-specific manner. TNF-α acts to reduce the abundance of CFTR mRNA post transcriptionaly in human colon cell lines (HT-29) and pulmonary cell lines (26, 27). Hull and Thomson (28) who studied 53 children with CF found that 20 of the patients had polymorphism in the promoter region of the TNF-α (-308A), which is known to be associated with higher levels of the cytokine. These children showed significantly lower FEV1 (forced expiratory volume per second) and poorer nutritional status than children without the polymorphism (28). A separate study of 180 Czech and Belgian CF patients aged 12-15 years, all of whom were homozygous for ΔF508, found that the 851C/T and -238G/A, but not -308G/A TNF-α polymorphisms were associated with the severity of lung disease (29). Recent studies have revealed that significant proportion of patients with symptoms of non-classic CF do not carry any CFTR gene mutation. However some mutations may have been missed and were not detected (23). Studies have shown that carriers of CFTR mutation are at higher risk of CF than the general population (30). Specific alleles of other genes or unfavorable environmental factors in combination with a single CFTR mutation could potentially produce a non-classic phenotype. Therefore, we first evaluated the potential modifying effect of the polymorphisms of genes other than CFTR on the progression and severity of CF, in patients with the same genotype, (∆F508/∆F508), with or without specific symptoms associated with lung and gastrointestinal function and weight loss. We compared the results obtained for the two groups based on the hypothesis that adverse and beneficial genetic variants would be enriched in these two groups of disease severity. We then compared those CF patients with non-classic phenotype who did not carry CFTR mutations with healthy control subjects based on the hypothesis that although the patients may not have mutations in the CFTR locus, cumulative effect of specific alleles of multiple CF modifier genes, such as TNF-α, may create the final phenotype. Alternatively, a combination of an undetected CFTR mutation and specific alleles of other genes (modifiers) may produce a non-classic phenotype. However, we did not find an association between the investigated polymorphic site and the non-classic CF disease or the severity of the classic form of the disease. Although we collected data from the maximum number of CF samples that were available during the study period, further studies and with larger sample size appear to be necessary for a more complete analysis of the probable associations between TNF-α or other modifier genes with CF in patients of this ethnic group. The results of the present study show that TNF-α gene does not act as a modifier in CF disease among the Iranian Azeri Turkish population.