1. Introduction
Allgrove or Achalasia-addisonianism-alacrimia syndrome (MIM: 231550) is caused by homozygous or compound heterozygous mutation in the gene encoding aladin (AAAS; 605378) on chromosome 12q13. Allgrove et al. (1978) described 2 pairs of siblings (aged 4 - 6 years) with the combination of the symptoms of achalasia, ACTH-resistant adrenal insufficiency and alacrima. This triad of symptoms is known as Allgrove syndrome (1, 2). Patients with Allgrove syndrome usually present dysphagia or severe hypoglycemic or hypotensive attacks, related to adrenal insufficiency in the first decade of life (3).
The onset of adrenal insufficiency is usually before puberty but it may become manifested in the third decade of life (4). In addition, progressive neurological abnormalities related to peripheral, central, or autonomic nervous system may be present (2) that is often associated with mental retardation (5). The phenotype of patients with Allgrove syndrome is highly variable (6).
This syndrome is caused by mutations of the AAAS gene, encoding a 546 amino acid protein ALADIN (alacrima- achalasia-adrenal insufficiency neurologic disorder). In Allgrove syndrome, the association of adrenal and neurologic disease is similar to X-linked adrenoleukodystrophy (7, 8).
In this report, we describe six Iranian patients with Allgrove syndrome, and discuss clinical and genetic features.
2. Methods
2.1. Patients and Samples
In this study we present 6 new patients of Allgrove syndrome. Patient 1 is a 12-year-old Iranian boy. He was a product of non-consanguineous marriage and born by natural delivery. He was confirmed to have alacrima at the age of 1 year. He developed adrenal insufficiency at the age of 11 years. Patient 2 is a 7-year-old Iranian girl, born to non-consanguineous parents. Alacrima was diagnosed at the age of 2 months. She was confirmed to have adrenal insufficiency at the age of 1 year and 6 months. Patient 3 is a 6-year-old Iranian boy, born to consanguineous parents. He was identified to have alacrima at the age of 1 year. He was confirmed to have adrenal insufficiency at the age of 2 years and 6 months. Patient 4 is a 6-year-old Iranian girl, born to non-consanguineous parents. Alacrima was diagnosed at the age of 6 months. She was confirmed to have adrenal insufficiency at the age of 4 years. Patient 5 is a 9-year-old Iranian girl. She was born to consanguineous parents. At the age of 1 year and 6 months she developed alacrima. Adrenal insufficiency was definitively confirmed at the age of 3 years. Patient 6 is an 8-year-old Iranian girl. She was born to non-consanguineous parents. She was diagnosed to have alacrima at the age of 8 months. She was confirmed to have adrenal insufficiency at the age of 5 years.
This study was approved by the ethics committee of the Medical University of Isfahan according to the national health and medical research council guidelines.
2.2. Molecular Analysis
DNA was extracted from whole-blood samples by genomic DNA isolation kit (Genet Bio, Korea). The exons 1 to 16 and some introns of the AAAS gene were amplified by the PCR. Primers used for PCR are listed in Table 1. PCR reactions were carried out in a total volume of 25 µL which contained 10 ng of DNA, 12.5 µL Taq DNA Polymerase (Master Mix(Ampliqon, Denmark) and 1 µL of each primer (10 pmol). After a brief centrifugation, the PCR plate was subjected to 35 cycles of the following conditions: (I) PCR activation at 94°C for 3 minutes, (II) denaturation at 94°C for 30 seconds, (III) annealing at 60°C for 30 seconds, (IV) extension at 72°C for 45 seconds, and (V) final extension at 72°C for 10 minutes. Presence of PCR products was evaluated on a 1.5% agarose gel, and then verified by complete nucleotide sequence analysis (Pishgam Biotech Co, Iran). After sequencing, alignment and analysis were carried out.
| Primer Name | Sequence |
|---|---|
| PF1 | CGCAGGAAGAAGCTTTGGAGG |
| PR1 | GTCTTAGCTCCCGTATCTGTGCAAC |
| PF2 | TTTGTGCTATAGTGGATCAATCTTCCTG |
| PR2 | CACATCATCTGAGGTCGGAAGTTC |
| PF3 | GGGATCAGAGTAAGCTGACCCAC |
| PR3 | GGAATGAGAATATGGTGAGCAATCAG |
| PF4 | AGGTGCAGGAACTCTTCATGTTAACC |
| PR4 | CGTAACCCACAGACACATTGTCC |
| PF5 | TGGGCCATCAGGATAGGAGG |
| PR5 | AGCAAATTACAGCTCAGGACCTCTAAG |
| PF6 | TTAGAGGTCCTGAGCTGTAATTTGCT |
| PR6 | GGGCACGGCCTCATTAGATTAAC |
| PF7 | CTTCCCAGTTAATCTAATGAGGC |
| PR7 | AAAGACAGACTGGTAACTGAGTGG |
List of the Primers Used in This Study
2.3. Bioinformatics Analysis
To in silico evaluate the possible effects of the identified variant on gene splicing, Human Splicing Finder (http://www.umd.be/HSF/, Marseille, France) and NetGene2 (http: //www.cbs.dtu.dk/services/NetGene2/, Lyngby, Denmark) softwares were used. Splice site scores were calculated using the ‘Berkeley Drosophila Genome Project (BDGP)’ software (www.fruitfly.org/seq_tools/splice.html).
3. Results
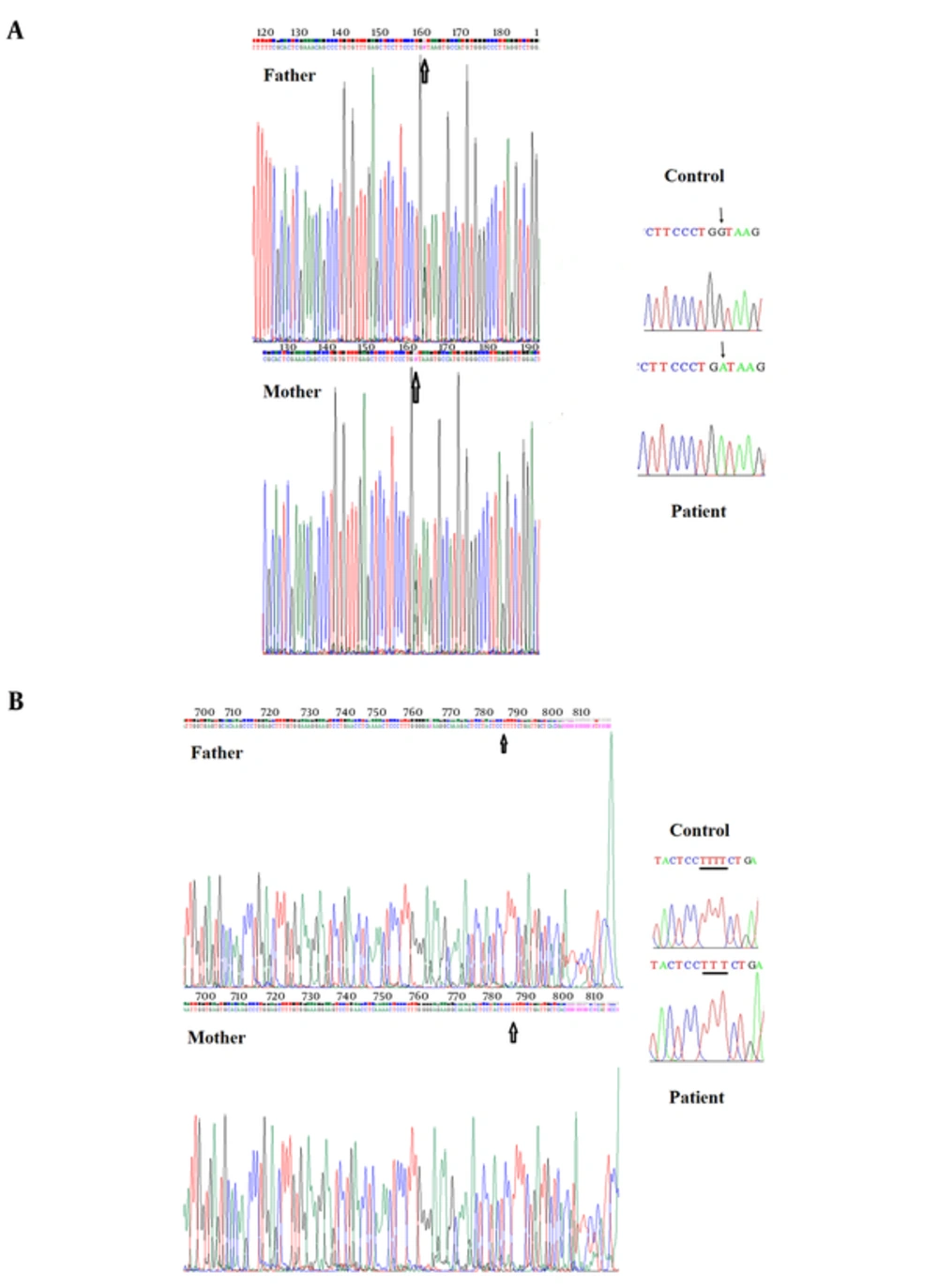
The sequencing results of all patients were analyzed and following mutations were found. We sequenced all 16 exons and some introns of the AAAS gene for the patient 1. The analysis of the AAAS gene of patient 1 demonstrated a G to A transition at the splice donor of exon 14 (IVS14 + 1G to A) (Figure 1A). The analysis of the AAAS gene for patient 2 revealed a novel mutation, including a T deletion in intron 5 (c.446 + 87del T) (Figure 1B). We analyzed sequence of all 16 exons and some introns of the AAAS gene for the patients 3 to 6, but no mutation was found. The results are summarized in Table 2.
A, DNA sequencing electropherogram depicting the IVS14 + 1 mutation. The arrow indicates site of mutation in the Patient 1, which is a G to A change. In the control, the arrow indicates the normal nucleotide. The parents are heterozygote, one allele is wild type and one allele is new mutation; B, DNA sequencing electropherogram depicting the c.446 + 87delT novel mutation. The underline indicates the site of mutation in Patient 2, which is a T nucleotide deletion. In the control, the underline indicates the normal nucleotide and the parents are normal.
| Patient No./Sex | Age | Manifestations | AAAS Gene |
|---|---|---|---|
| 1/M | 12 | Alacrima, Adrenal insufficiency | IVS14 + 1 G > A |
| 2/F | 7 | Alacrima, Adrenal insufficiency | c.446 + 87del T |
| 3/M | 6 | Alacrima, Adrenal insufficiency | Normal |
| 4/F | 6 | Alacrima, Adrenal insufficiency | Normal |
| 5/F | 9 | Alacrima, Adrenal insufficiency | Normal |
| 6/F | 8 | Alacrima, Adrenal insufficiency | Normal |
Summary of Patients Information
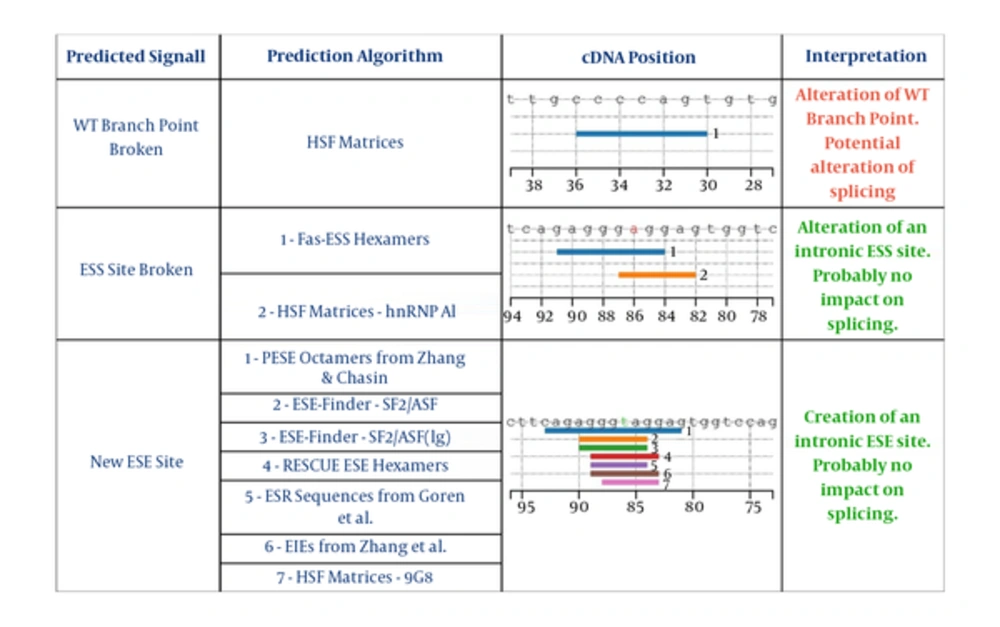
To determine whether the AAAS c.446 + 87delT variant is pathogenic, we performed bioinformatics predictions using specific tools. This in silico analysis proposed alteration of splicing reactions that probably makes the retention of AAAS intron 5 (Table 3). Especially, the Human Splicing Finder results show breakage of the wild type (wt) donor site. The variant seems to generate different potential Exonic Splicing Enhancers (ESEs) from position +82 to +94, and also the potential alteration of one of these at the position. All these molecular events probably create an alternative splicing process (Figure 2).
| Splice-Site Analysis Tools | AAAS, c.446 + 87del T |
|---|---|
| Human Splicing Finder software | √ WT branch point broken: Alteration of WT Branch Point |
| Potential alteration of splicing | |
| √ ESS Site broken: Alteration of an intronic ESS site. | |
| Probably no impact on splicing. | |
| √ New ESE Site: Creation of an intronic ESE site. | |
| Probably no impact on splicing. | |
| Berkely Drosophila Genome Project (BDGP) | Breakage of the wild type (wt) donor site (score 0.4) |
| NetGene 2 (NG2) |
Splice Site Analysis of the Intron Variant Mutation c.446 + 87del T in AAAS Gene
4. Discussion
Since first described by Allgrove et al. in 1978 (1), many patients with Allgrove syndrome have been reported worldwide. Studies have identified mutations in the AAAS gene that are responsible for this disease; therefore, nowadays molecular analysis is usually accomplished to confirm the clinical diagnosis. Mutations in the AAAS gene have been described in several individuals (Table 4). The AAAS gene consists of 16 exons and is translated to a 546 amino acid protein called ALADIN (alacrima-achalasia-adrenal insufficiency neurologic disorder). ALADIN does not show major homology to any known protein but has four WD-repeats (tryptophan-aspartate repeat). WD proteins are involved in a variety of cellular processes such as cell cycle progression, cell fate determination, signal transduction, gene regulation, intracellular trafficking, transcription and apoptosis (9, 10). The function of ALADIN protein is still unknown, but proteomic analysis has shown that ALADIN is a part of nuclear pore complex (NPC) (11).
| Location | cDNA Mutation | Predicted Effect | References |
|---|---|---|---|
| Ex 1 | c.43C > A | p.Q15K | (6, 7, 12, 13) |
| Ex 2 | c.210delC | p.170fsX92 | (13) |
| Ex 2 | c.211delC | p.H71fsX92 | (14) |
| Ex 2 | c.251G > A | p.W84X | (7) |
| Ex 3 | c.352delT | p.F90fs | (15) |
| Ex 4 | c.355C > T | p.R119X | (14) |
| IVS 4 | IVS4-2A > G | (8) | |
| Ex 5 | c.433C > T | p.Q145X | (14) |
| In5 | c.446 + 87del T | This report | |
| IVS 5 | IVS5 + 3insT | (14) | |
| Ex 6 | c.518A > T | p. D173V | (15) |
| Ex 6 | c.479A > G | p.H160R | (7) |
| Ex 6 | c.470-471delTT | p.F157fsX171 | (7) |
| Ex 7 | c.580C > T | p.R194X | (16, 17) |
| Ex 7 | c.678C > T | p.R230X | (13, 14) |
| Ex 8 | c.787T > C | p.S263P | (6, 7, 13) |
| IVS8 | IVS8 + 1G > A | (18) | |
| Ex 9 | c.856C > T | p.R286X | (7, 19) |
| Ex 9 | c.928-931delGTCT | p.V310FfsX42 | (15) |
| Ex 9 | c.934C > T | p.R312X | (8) |
| Ex 9 | c.991T > C | p.C331R | (15) |
| Ex 10 | c.981-982insT | p.S328fsX363 | (8) |
| Ex 10 | c.1238T > C | p.V313A | (13) |
| Ex 11 | c.1024C > T | p.R342X | (7) |
| Ex 11 | c.1066-1067delCT | p.L356fsX362 | (13) |
| IVS 11 | IVS11 + 1G > A | (12) | |
| Ex 12 | c.1104-1105insC | p.D368fsX382 | (13) |
| Ex 12 | c.1144-1147delTCTG | p.S382fsX413 | (13) |
| Ex 12 | c.1159C > T | p.Q387X | (6) |
| Ex 13 | c.1191insA | p.397fxs27 | (13) |
| Ex 14 | c.1374-1176delTTCinsA | p.F431X | (15) |
| IVS14 | IVS14 + 1G > A | (8, 12, 14, 20) | |
| Ex 15 | c.1366C > T | p.Q456X | (14) |
| Ex 15 | c.1368-1372delGCTCA | p.X492 | (19) |
| Ex 15 | c.1389delC | p.S463fsX549 | (7) |
| Ex 16 | c.1421G > A | p.W474X | (13) |
| Ex 16 | c.1422G > C | p.W474C | (15) |
| Ex 16 | c.1432C > T | p.R487X | (8, 20, 21) |
List of Reported Mutations in Patients with Allgrove Syndrome
Tullio-Pelet et al (2000) found 5 homozygous truncating mutations in the AAAS gene in unrelated patients with triple-A syndrome. They described the founder effect to a single splice donor mutation that happened more than 2,400 years ago in North African families (8). Sandrini et al (2001) investigated 6 families with Allgrove syndrome and 4 with isolated ACTH resistance. Four triple-A syndrome families were from Puerto Rico and most of the remaining 6 families were Caucasian families from North America. All of the triple-A syndrome families, showed mutations in AAAS gene, but no kindred with ACTH resistance. Apparently due to a founder effect, the IVS14 + 1G-A donor splice mutation was found in all Puerto Rican families. In addition, a North American kindred was heterozygous for this mutation. One Puerto Rican family had a new splice donor site mutation in exon 11 of the AAAS gene, IVS11 + 1G-A; the proband was a compound heterozygote. In a Canadian triple-A syndrome kindred, a gln15-to-lys point mutation in homozygote state was determined with a milder phenotype. The patients with the same AAAS defect showed significant clinical variability (12). Handschug et al identified 8 different homozygous and compound heterozygous mutations in the AAAS gene in 9 patients with Allgrove syndrome. Most of these mutations cause truncation of protein (7, 22).
In one patient, we identified a IVS14 + 1 G > A mutation, which has been previously reported in Algerian and Hispanic patients and, hence, is highly expected to be pathogenic (8, 12). This splice site mutation produces abnormal transcripts which cause premature termination of the predicted protein (8). In 4 patients, we couldn’t detect any mutation. We determined a new mutation (c.446 + 87del T) in the AAAS gene in a patient. We speculate that this deletion causes splicing defect in intron 5 which results in a premature termination and non-functional ALADIN protein. However, to define whether it affects AAAS gene expression or function, more detailed studies will be required. In conclusion, the most frequent mutations were not detected in our patients and perhaps further investigation in AAA gene is required to find all mutations in Iran. Since the molecular genetic testing results may influence the therapy and prognosis of Allgrove patients, this paper contributes to understanding the molecular basis of Allgrove in Iranian patients.