1. Background
Herpes viruses are classified into three types including herpes simplex virus-1 (HSV-1), herpes simplex virus-2 (HSV-2), varicella zoster virus (VZV), Epstein-Barr virus (EBV), cytomegalovirus (CMV), and human herpes virus 6 (HHV-6A, HHV-6B). They can cause various diseases ranging from herpes to encephalitis, hearing loss or pediatric lymphomas (1-3). Furthermore, human herpes viruses have a tendency to be latent and may be reactivated under some conditions such as weakened immunity. The reactivation of latent infections may lead to serious consequences such as postherpetic neuralgia, encephalitis, and EBV associated tumors (4-6). Moreover, many children in the world are infected with herpes viruses (5, 7, 8). Thus, it is important to rapidly and simultaneously identify all these seven human herpes viruses in blood samples and provide the patients with timely treatment.
However, herpesvirus infections are often non-specific and cannot be distinguished from other viral infections. At present, virus isolation and serological tests are common methods for diagnosis of herpesvirus infections, although both of them are slow, labor-intensive, and inaccurate. Cross-reactivity can also impede the clinical application of immunological assays (9, 10). Moreover, PCR method is limited to detect only one type of pathogens in spite of high sensitivity and specificity (11). Therefore, in clinical work, it would be better to rapidly and simultaneously detect the pathogens and identify the type of herpes viruses.
2. Objectives
In this study, we used multiplex PCR-based DNA microarray technology, which can offer the high-throughput results with high sensitivity and specificity, to detect and identify the type of herpes viruses rapidly and simultaneously in blood samples (12).
3. Methods
3.1. Patients and Virus Strains
A total of 108 blood specimens from inpatients who were diagnosed with viral infection in our hospital were collected from January 2015 to January 2016 in Hangzhou city, Zhejiang province. Their ages ranged from 1 year to 10 years. The inclusion criteria included 1) presence of fever (body temperature ≥ 38°C), chills, vomit, headache, anorexia; and 2) blood test result showing an increase in the lymphocyte count. The exclusion criteria included children infected with bacteria, fungi, or mycoplasma detected by PCR method, fungal culture method, or serological tests. HSV-1, HSV-2, EBV, VZV, HCMV, HHV-6A, and HHV-6B strains were conserved in our hospital. The negative controls were hepatitis B virus (HBV), U. Urealyticum, M. pneumonia, S. aureus, and E. coli strains.
This study was approved by our hospital and informed consent was obtained.
3.2. Multiplex PCR-Based DNA Microarray Technology
200 μL of EDTA anticoagulant blood samples were collected from each infected child. Total DNA was extracted by using Axyprep Blood Genomic DNA Miniprep Kit (Axygen, Shanghai, China). This technology has been described previously in detail (13, 14). First, multiplex PCR was performed. The primers designed in the highly conserved regions of human herpes viruses’ genes and probes targeted at the sequences of human herpes viruses are shown in Table 1. PCR conditions were as follows: 95°C for 5 minutes and 35 cycles at 95°C for 30 seconds, 58°C for 30 seconds and 72°C for 30 seconds; final extension at 72°C for 5 minutes. PCR products were examined by electrophoresis on 0.8% formaldehyde/agarose gel in 1 × borate buffer. Second, DNA microarray was prepared, hybridized, and scanned (14). Finally, the data were analyzed with ScanArray Express software 2.0. The positive criterion was considered an Is/b value of greater than or equal to 3. The Is/b was defined as the ratio of the mean spot intensity to the mean background intensity.
| Sequences (5’ - 3’) | Target Gene of Virus | Size of Product, bp | |
|---|---|---|---|
| Primers 1 | Cy5-TCATCTACGGGGACACGGAC | HSV-1 UL30 | 232 |
| CGCACCAGATCCACGCCCTT | HSV-2 UL30 | 232 | |
| HCMV UL54 | 235 | ||
| Primers 2 | Cy5-TCATCTACGGGGACACGGAC | EBV BALF5 | 227 |
| GAGCTCCACCCCCTTCATC | |||
| Primers 3 | CY5-TAGGCGGATGGTGGACGACT | VZV ORF28 | 224 |
| GCCCGCAAACTTGTAGAACTG | |||
| Primers 4 | Cy5-CCGTGAAGTTGGGGGATGAG | HHV-6A UL38 | 252 |
| CAGAAGCAGCAATCGCAACAC | HHV-6B UL38 | 252 | |
| Probes 1 | GGCACAGCACAAAGATGGA | HSV-1 UL30 | |
| Probes 2 | GGCACAAAACGAAAATGGA | HSV-2 UL30 | |
| Probes 3 | ACGAAAGCGGACAAACACG | HCMV UL54 | |
| Probes 4 | CGGCACTCGATAAACAGCG | EBV BALF5 | |
| Probes 5 | GTTTGTTATGACGGCCGAG | VZV ORF28 | |
| Probes 6 | AGACTCACGGATACAAGGC | HHV-6A U38 | |
| Probes 7 | AGACTCGCGAACACAAGGC | HHV-6B U38 | |
| probe for orientation | GTCCGTGTCCCCGTAGATGA |
aPrimers 1 were used to simultaneously amplify HSV-1, HSV-2 and CMV, while Primers 4 were applied for both HHV-6A and HHV-6B.
3.3. PCR Method
TaqMan PCR kits (Daan gene, Guangdong, China) were used to detect human herpes viruses. We followed the recommended instructions and performed reactions on a Sequence Detection System GeneAmp5700 (PerkinElmer, Massachusetts, USA). According to our previous study, the conditions were: 93°C for 2 minutes; 10 cycles at 93°C for 45 seconds and 55°C for 60 seconds; and 30 cycles at 93°C for 30 seconds and 55°C for 45 seconds (13).
3.4. IgM ELISA
Virus-specific IgM antibody was determined by ELISA kits (Euroimmun, Luebeck, Germany). The process was as follows: at first, diluted samples and virus strains were incubated in microplates and reacted with anti-human herpes viruses IgM for 30 minutes at room temperature. Then, the plates were incubated with diluted streptavidin–peroxidase conjugate for 30 minutes after washing. Subsequently, the plates were washed again and the substrate was added (TMB/H2O2), followed by 15 minutes incubation. Finally, the stopping solution (0.5 M sulfuric acid) was added and the color intensity measured at 450 nm.
4. Results
4.1. Specificity and Sensitivity of the DNA Microarray Technology
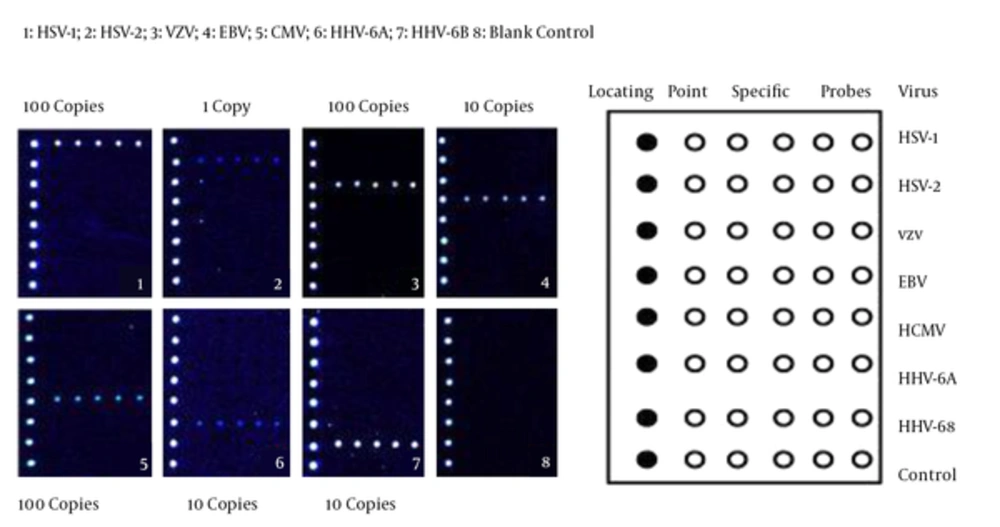
To evaluate the specificity of multiplex PCR-based DNA microarray technology, DNAs were isolated from the virus strains and controls, amplified by multiplex PCR, and tested by DNA microarray technology. As shown in Figure 1, these seven common human herpes viruses were all positive by detecting the hybridization signals, while no signals were found in other virus strains. Moreover, no fluorescence signals were detected and no cross-reactions were observed in the control group (composed of S. aureus, E. coli, HBV, U. Urealyticum and M. pneumonia).
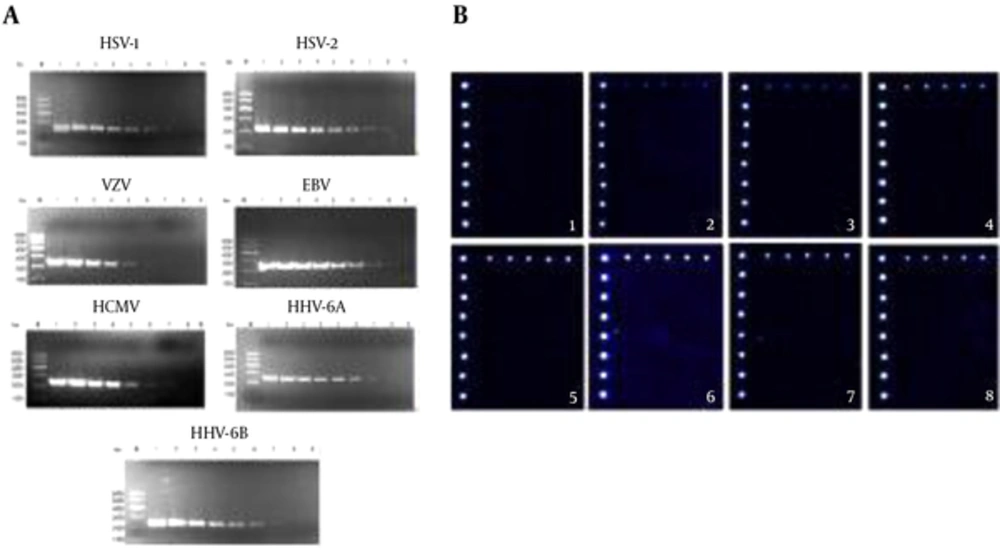
To determine the sensitivity of this technology, ten-fold serially diluted templates of seven human herpes viruses ranging from 100 to 107 copies/μL were subjected to multiplex PCR with specific primers for herpes viruses and examined by electrophoresis on 0.8% formaldehyde/agarose gel in 1 × borate buffer. As shown in Figure 2A, the results suggested that the detection limits were 102 copies/μL for VZV and CMV, 101 copies/μL for HSV-1, EBV, HHV-6A and HHV-6B, and 100 copies/μL for HSV-2.
A, the sensitivity of detection by multiplex PCR method. M, marker; 1, 107 copy/mL; 2, 106 copy/mL; 3, 105 copy/mL; 4, 104 copy/mL; 5, 103 copy/mL; 6, 102 copy/mL; 7, 101 copy/mL; 8, 100 copy/mL; 9, blank control; B, the sensitivity of detection by DNA microarray technology. 1, 100 copy/mL for HSV-1; 2, 101 copy/mL for HSV-1; 3, 102 copy/mL for HSV-1; 4, 103 copy/mL for HSV-1; 5, 104 copy/mL for HSV-1; 6, 105 copy/mL for HSV-1; 7, 106 copy/mL for HSV-1; 8, 107 copy/mL for HSV-1.
Furthermore, signals from all five detection spots in each line were collected, averaged and compared with the mean background intensity. An Is/b value of equal to or greater than 3 indicated the positive signal. The detection limit was 101 copies/μL for HSV-1 which was in line with DNA electrophoresis (Figure 2B).
4.2. Comparison of DNA Microarray Technology, PCR Method and IgM ELISA
Out of 108 blood specimens, 16 were tested positive by multiplex PCR-based DNA microarray technology including 6 EBV, 5 HCMV, 2 HHV-6B, 1 HSV-2, 1 VZV, and 1 both EBV and CMV (Table 2). 75% of the positive samples were EBV and HCMV. The sensitivity and specificity of multiplex PCR-based DNA microarray technology for detecting human herpes viruses were 100% when compared to the PCR method. None of 30 specimens in the control group were detected positive by both multiplex PCR-based DNA microarray technology and PCR method. Then, these 108 blood specimens were examined by IgM ELISA. Out of 16 positive specimens, 11 specimens were positive, and 2 specimens were negative. The rest 3 specimens although were positive in IgM ELISA, were negative in multiplex PCR-based DNA microarray technology. Furthermore, the negative samples confirmed by multiplex PCR-based DNA microarray technology and PCR method were also negative in IgM ELISA. Thus, the positive and negative rates of IgM ELISA in accordance with multiplex PCR-based DNA microarray technology were 68.8 % and 100%, respectively.
| DNA Microarray Results | PCR Results | IgM -ELISA Results |
|---|---|---|
| 6 for EBV | 6 for EBV | 5 for EBV; 1 for HCMV |
| 5 for HCMV | 5 for HCMV | 3 for HCMV; 1 for HSV-2 and EBV; 1 for negative |
| 1 for HSV-2 | 1 for HSV-2 | 1 for HSV-2 |
| 1 for VZV | 1 for VZV | 1 for negative |
| 2 for HHV-6B | 2 for HHV-6B | 1 for HSV-2; 1 for HHV-6B |
| 1 for EBV and HCMV | 1 for EBV and HCMV | 1 for EBV and HCMV |
5. Discussion
Human herpes viruses not only can cause acute infections, but also may be reactivated from the status of latent infection (15). They can also affect the central nervous system and organs, leading to encephalitis, hepatitis, and Hodgkin lymphoma (2, 16). Among herpes viruses, EBV and CMV are more prevalent in the world and are commonly contracted during infancy (17, 18). EBV and CMV reside in B lymphocytes and myeloid progenitor cells, respectively. While primary EBV and HCMV infections are mild or asymptomatic, reactivation of latency can cause fatal diseases. EBV has infected more than 90% of individuals around the world and can establish the latency after primary infection (17). Exceptionally, primary infection with EBV in older children can cause acute infectious mononucleosis instead of establishment of latency (18). Moreover, the expression of latency-related proteins and miRNA has been considered to play a role in the pathogenesis of different lymphomas (19). Serological tests are used in clinics to diagnose EBV, but heterophile antibodies are non-specific and not produced in some patients (17). HCMV is the major cause of congenital infection, affecting 0.3% to 2.3% of newborns, that can lead to severe neurological sequelae such as hearing loss and congenital defects (20, 21). It also infects the immunocompromised individuals causing fatal pneumonia and colitis. What’s more, in transplant recipients, HCMV infection not only increases the risk of graft rejection, but also rises the treatment costs (21). Thus, early antiviral interventions can help the body eliminate the pathogens and prevent from establishing latent infections as well as playing an important role in the prevention of CNS infections and pediatric lymphomas. Therefore, timely diagnosis is a vital need.
In this study, PCR-based DNA microarray technology was used to diagnose herpes viruses in blood samples. The sensitivity and specificity of multiplex PCR-based microarray technology were 100% when compared to PCR method. The results also showed that crude agreement, positive rate and negative rate of IgM ELISA in accordance with multiplex PCR-based DNA microarray technology were 95.4%, 68.8%, and 100%, respectively. But there were some differences between PCR-based DNA microarray method and IgM ELISA. The differences may be explained as follows: First, IgM may exist in blood due to non-specific reactions of other pathogens. For example, some patients infected with EBV do not produce these antibodies or may produce non-specific antibodies. Second, it is well known that the results of serological tests can be affected dramatically by the immunological state of patients such as in immunosuppressed populations. As we all know, herpes viruses, especially HCMV and EBV, often infect the immunosuppressed individuals who may not produce antibodies. Third, IgM antibody usually persists for about 1 month, and the viral load of these 3 positive samples were low at about 1000 copies/μL. Perhaps, these 3 children with different results had been infected with other viruses before (data not shown).
Among 108 samples, one blood sample was found co-infected with EBV and HCMV detected by PCR-based DNA microarray technology that was also confirmed by PCR method. It indicated that this method can detect mixed infections of human herpes viruses simultaneously. But it is more complicated and time-consuming than PCR method to detect one type of herpes viruses. Moreover, viral DNA load cannot be detected using this method. Therefore, in clinical work, whether the antiviral treatment is effective or not can be independent of the results of PCR-based DNA microarray technology due to its inability to detect viral load.
In conclusion, multiplex PCR-based DNA microarray technology can provide high-throughput results with low false negative rates for simultaneous and rapid identification of all the seven common human herpes viruses in blood specimens from children with viral infections. The crude agreement between IgM ELISA and multiplex PCR-based DNA microarray was 95.4% which indicated that multiplex PCR-based DNA microarray technology may someday be an alternative for IgM ELISA.