1. Background
LPS-responsive beige-like anchor protein (LRBA) deficiency is a rare inherited common variable immunodeficiency (CVID) caused by loss-of-function mutations in LRBA gene. Affected individuals were usually reported to suffer from immune deficiency, lymphoproliferation and various organ-specific autoimmunity, which severely affected the patients’ physical health and imposes a heavy economic burden on the patients (1-5).
LRBA gene, which is located on chromosome 4q31.3, contains 57 exons and encodes a large protein containing 2851 amino acid residues. LRBA belongs to the BEACH-WD40 protein family composed of multiple functional domains, including Concanavalin A (ConA)-like lectin binding domain, vacuolar protein sorting-27, hepatocyte growth factor-regulated tyrosine kinase substrate domain and signal transducing adaptor molecule (VHS) domain, two PKA regulatory subunit (RII) binding motifs, WD-like (WDL), and Beige and Chediak-Higashi (BEACH) domains, plekstrin homology (PH)-like domain and five WD40 repeats (6). LRBA is broadly expressed in several human tissues and involved in intracellular vesicle trafficking and signal transduction. Besides, LRBA deficiency was reported to be associated with a diminished expression of CTLA4 on the surface of Treg cells (7).
LRBA gene was firstly identified as an CVID-causing gene in 2012. Three papers were published almost at the same time to categorize LRBA deficiency as a common variable immunodeficiency (CVID), with autoimmunity and inflammation (1, 8, 9). However, its clinical spectrum has been extended with the increasing publications worldwide in the recent years, which included recurrent infections, hypogammaglobulinemia, inflammatory bowel disease, enteropathy, splenomegaly, hepatomegaly, lymphadenopathy, growth retardation, atopic skin disease, EBV infections and so on (1-4, 10). Immunologic testing showed that a majority of LRBA deficient patients had reduced total B cells and B cell subsets, as well as Treg cells (3, 10, 11). However, the broad phenotypic spectrum made the clinical diagnosis of LRBA deficiency difficult, which made genetic analysis to distinguish it from other diseases necessary.
2. Objectives
Herein, we describe a Chinese patient with compound heterozygous mutations in LRBA gene, who had decreased B cells, but extremely high level of IgE, mainly suffering from recurrent respiratory tract infections, systemic vasculitis and EBV infection. To our knowledge, this is the first Chinese LRBA deficient patient reported.
3. Methods
3.1. Patient
The affected patient with LRBA deficiency was enrolled in Shanghai Children’s Medical Center, China. Healthy control children were age matched. All study participants were recruited with written informed consent approved by the Ethics Committee of Shanghai Children’s Medical Center.
3.2. Targeted Gene Capture and Next-Generation Sequencing
DNA library was prepared according to illumine standard protocols, then captured with an immunodeficiency related gene panel, which included all PID-causing genes published in the International Union of Immunological Societies Expert Committee for Primary Immunodeficiency on 2015, by using biotinylated oligo-probes. The capture experiment was conducted according to manufacturer’s protocol (MyGenostics, Beijing, China) as previously described (12). In brief, 1μg DNA library was mixed with Buffer BL and probes, incubated at 95°C for 7 min and 65°C for 2 min. Then, 65°C Buffer HY was added to the mixture, and incubated at 65°C for 22 hours for hybridization. Then MyOne beads (Life Technology) was used to bind the biotinylated oligo-probes. The bound DNA was eluted and amplified through PCR, and then purified using SPRI beads (Beckman Coulter). Finally, Illumina HiSeq 2000 sequencing machine was used to sequence the libraries.
3.3. Bioinformatics Analysis
High-quality reads were retrieved through eliminating low quality reads and adaptor sequences, then aligned with the human reference genome (hg19) using SOAPaligner program. The SNPs was firstly identified using the SOAPsnp program after PCR duplicates were removed by the Picard software. Then, the reads were realigned to the reference genome to detect insertions or deletions by using the GATK program. The identified SNPs and InDels were annotated by using Exome-assistant program. MagicViewer was then used to view the short read alignment and validate the candidate SNPs and InDels.
3.4. PCR and Sanger Sequencing
Genomic DNA was isolated from peripheral blood by using RelaxGene blood DNA isolation kit (Tiangen Biotech Co. Ltd.) according to the manufacturer's instructions. PCR Products sent for Sanger sequencing. Sequence analysis software (Gene Runner) was used to identify gene mutations.
3.5. B Cell Immunophenotyping
B cell subsets were classified by flow cytometry according to the expression of surface markers as follows: hematopoietic stem cell (HSC): CD34+CD10-CD19-IgM-; common lymphoid progenitor (CLP): CD34+CD10+CD19-IgM-; Pro-B cells: CD34+CD10+CD19+IgM-; Pre-B cells: CD34-CD10+CD19+IgM-; immature B cells: CD19+CD10+CD27-CD38+; memory B cells: CD19+CD27; naïve B cells: CD19+CD27-IgD+; non-switched memory B cells: CD19+CD27+IgD+; switched memory B cells : CD19+CD27+IgD-; double negative B cells: CD19+CD27-IgD-; plasma cells: CD19+CD10-CD27++CD38++; naïve B1: CD20+CD27-CD43-; memory B1: CD20+CD27+CD43-; B1: CD20+CD27+CD43+.
4. Results
4.1. Clinical Presentations
In this study, the male patient, who was born in 2005, was the first child from non-consanguineous parents and there was no history of similar disorders in their families. At 2 years of age, he got fever without obvious cause (maximum temperature, 39.6°C), paroxysmal and non-spasmodic cough and wheeze, and then was hospitalized with asthmatic suffocating pneumonia in a local hospital. After two months of treatment, His cough and wheeze was relieved, but he still had fever. In addition, enlarged cervical lymph nodes were observed in the patient.
The patient went to Beijing children’s hospital to seek treatment when 3 years old. At the time of admission, general physical examination showed that he had congested throat and enlarged tonsils (I degree). Some enlarged lymph nodes were also observed in his neck and groin bilaterally, which were soft, painful to touch and easy to move. Laboratory testing results showed that he had normal peripheral blood leukocytes, but raised serum C-reactive protein (CRP) levels and erythrocyte sedimentation rates (ESR), as well as positive Epstein-Barr virus (EBV) capsid antigen (VCA) IgG and EB VCA IgA, thus leading to a diagnosis of EBV-induced infectious mononucleosis complicated by bacterial infection. Anti-infective agents including ganciclovir and cefotiam were given to the patient to treat infections. During hospitalization, a progressive dilatation of coronary artery was observed in this patient by cardiac color ultrasound, while no obvious endangium lesions were detected by vascular ultrasound. Combined with his recurrent fever, relapse of raised ESR and CRP, positive lupus anticoagulant test, the patient was given the diagnosis of systemic vasculitis. Therefore, he was given a treatment with aspirin, dipyridamole and intravenous immunoglobulin. Despite these aggressive treatments, relapsing unexplained fever still occurred in this patient. Prednisone was administerd to reduce his fever. After resuming normal body temperature, he was discharged from the hospital.
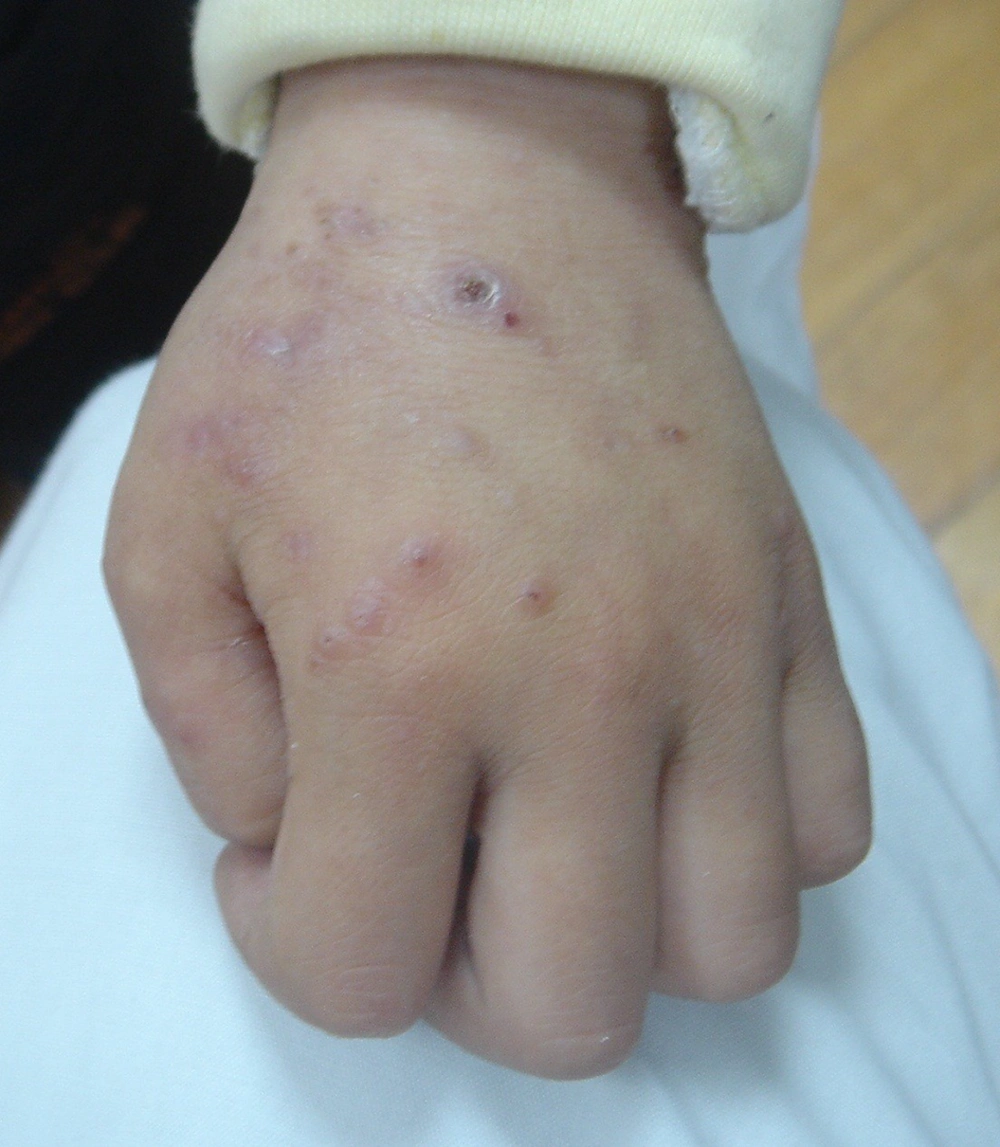
After discharge, he still had a recurrent fever. Vaccinia-like vesicles were also observed on his face and limbs, rupture of which led to ulceration (Figure 1). In November 2008, he came to our hospital to seek treatment. A clinical workup showed his body temperature was 39°C. Enlarged lymph nodes were observed in his ears, jaw, neck and right groin. Besides, he was reported to have swollen lip with obvious bleeding point and hepatosplenomegaly (at 1 cm under the ribs). Immunological tests showed that his serum IgG, IgA and IgM level were normal, while his serum IgE was up to 29400 IU/mL. Biopsies of skin vesicles showed allergic inflammation in the cutaneous and subcutaneous tissue, accompanied by focal necrosis and infiltration of eosinophils. Granulomatous lymphadenitis was also found in his right groin. The patient was suspected to have Hyper-IgE syndrome, but there were no STAT3 and DOCK8 mutations found.
After discharge, he had less fever, but lymph nodes enlargement and hepatosplenomegaly were still observed. Besides, he suffered from nasosinusitis and oral candidiasis, saprodontia, tooth loss, poor appetite and hard stools. In November 2014, he was hospitalized again because of fever. A routine blood examination showed he had decreased white blood cell (WBC), polymorphonuclear cell (PMN), lymphocyte (LYM) counts and hemoglobin (HGB). Vancomycin and fosfomycin was used to combat the infection, but the treatment was ineffective. The patient still had fever, and gradually developed severe hepatosplenomegaly, digestive hemorrhage, vomiting, hematochezia and dyspnea. Suspended red blood cells were infused to correct anemia, and omeprazole acetic acid was used for symptomatic treatment. However, these treatments were ineffective, and he died of multiple organ failure as a 9-year old in September, 2014.
4.2. Laboratory Analysis
The patient’s peripheral blood immunological data is shown in Table 1. He had normal lymphocytes and T cell subsets, but very low percentage of CD19+B lymphocytes. Interesting is that despite the low percentage of B cells, he was reported to have normal level of serum IgG, IgA, IgM, but extremely high level of IgE.
Neutrophil oxidative burst test was performed in this patient. The mean dihydrorhodamine 123 (DHR) stimulation index (SI) indicating the residual reactive oxygen intermediates (ROI) production capacity of neutrophils was 41.2 in the patient; whereas his parents’ results were 140.97 and 160.21, respectively (Table 1). These results suggested that the patient had defective NADPH oxidase activity.
| Variables | Values | ||
|---|---|---|---|
| Lymphocyte counts, 109/L | |||
| Age at measurement, y | 3.3 | 3.4 | |
| 2.55 (1.2 - 3.4) | 3.2 (1.2 - 3.4) | ||
| T lymphocyte cell subsets | |||
| Age at measurement, y | 3.3 | 3.4 | |
| CD3+ T cell counts, 109/L | 1.9 (0.7 - 2.10) | 2.1 (0.7 - 2.10) | |
| CD3+, % | 73.30 (64.19 - 75.77) | 68.60 (64.19 - 75.77) | |
| CD3+/CD4+, % | 46.10 (30.09 - 40.41)↑ | 41.60 (30.09 - 40.41)↑ | |
| CD3+/CD8+, % | 25.70 (20.74 - 29.42) | 23.20 (20.74 - 29.42) | |
| CD4+/CD8+, % | 1.79 | 1.79 | |
| B lymphocyte cell subsets | |||
| Group name | Patient | Age matched controls | |
| CD19+ B cell counts, 109/L | 0.03↓ | 0.12 - 0.56a | |
| CD19+, % | 1.132↓ | 4.3 - 18.2a | |
| Immunoglobulin levels | |||
| Age at measurement, y | 3.6 | 3.9 | 5.8 |
| IgG, g/L | 18.7 (6.94 - 16.85)↑ | 16.2 (6.94 - 16.85) | 16.40 (7.51 - 15.60)↑ |
| IgM, g/L | 1.09 (0.60 - 2.77) | 1.95 (0.60 - 2.77) | 1.49 (0.46 - 3.04) |
| IgA, g/L | 2.39 (0.68 - 3.82) | 2.54 (0.68 - 3.82) | 6.05 (0.82 - 4.53)↑ |
| IgE, IU/mL | 8600 (5.00 - 165.3)↑ | 5220 (5.00 - 165.3)↑ | 29400 (5.00 - 165.3)↑ |
| Neutrophil oxidative burst test, SI | |||
| Patient | 41.2 | Normal range | |
| Father | 140.97 | 127.9 (85.2 - 264.6) | |
| Mother | 160.21 | ||
Laboratory Parameters of the Patient with LRBA Gene Mutations
4.3. Mutation Analysis
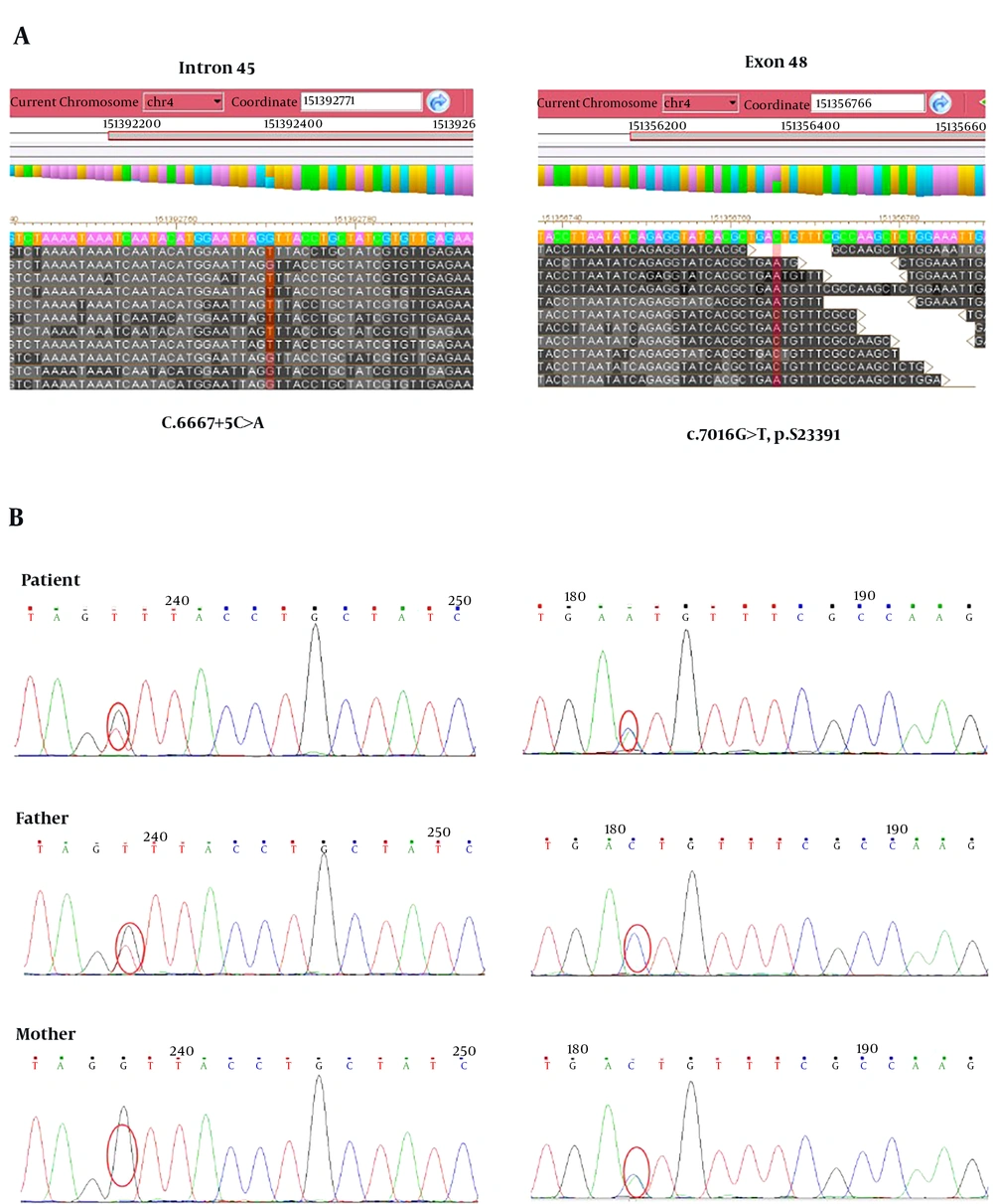
To find out the disease causing gene, targeted gene capture combined with next-generation sequencing technology was applied in this study. The results showed that this patient was a compound heterozygote for novel mutations consisting of a C to T transition at position 7016 of exon 46 (c.7016G>T, p.S2339I) and a C to A transition at the fifth nucleotide of intron 45 (c.6667+5C>A, splicing) in LRBA gene (Figure 2A). The frequency of the T variant at c.7016G>T has been reported to be 4.6% in 1000 genomes, and there was no population frequency data of c.6667+5C>A. The following Sanger sequencing results showed that the compound heterozygous mutations were inherited from his parents, in which his father carried a heterozygous c.7016G>T mutation and his mother had a heterozygous c.6667+5C>A mutation (Figure 1B). Mutation damage prediction was made using standard open access software. The p.S2350I was predicted to be probably damaging with a score of 0.963 by using PolyPhen2 (http://genetics.bwh.harvard.edu/pph2); disease causing by using MutationTaster (http://www.mutationtaster.org); affecting protein function with a score of 0.00 through Sorting Intolerant from Tolerant (SIFT; http://sift.jcvi.org). The c.6667+5C>A was predicted to be disease causing by using MutationTaster.
Mutation analysis of LRBA deficiency. A, Targeted gene capture followed by next-generation sequencing technology was used to screen mutations causing disease in the patient. B, Verify LRBA gene mutations in the patient and his parents by Sanger sequencing. Compound heterozygous mutations including c.6667+5C>A and c.7016G>T were found in the patient. A heterozygous mutation of c.6667+5C>A and c.7016G>T was found in his father and his mother, respectively.
4.4. B Cell Immunophenotyping
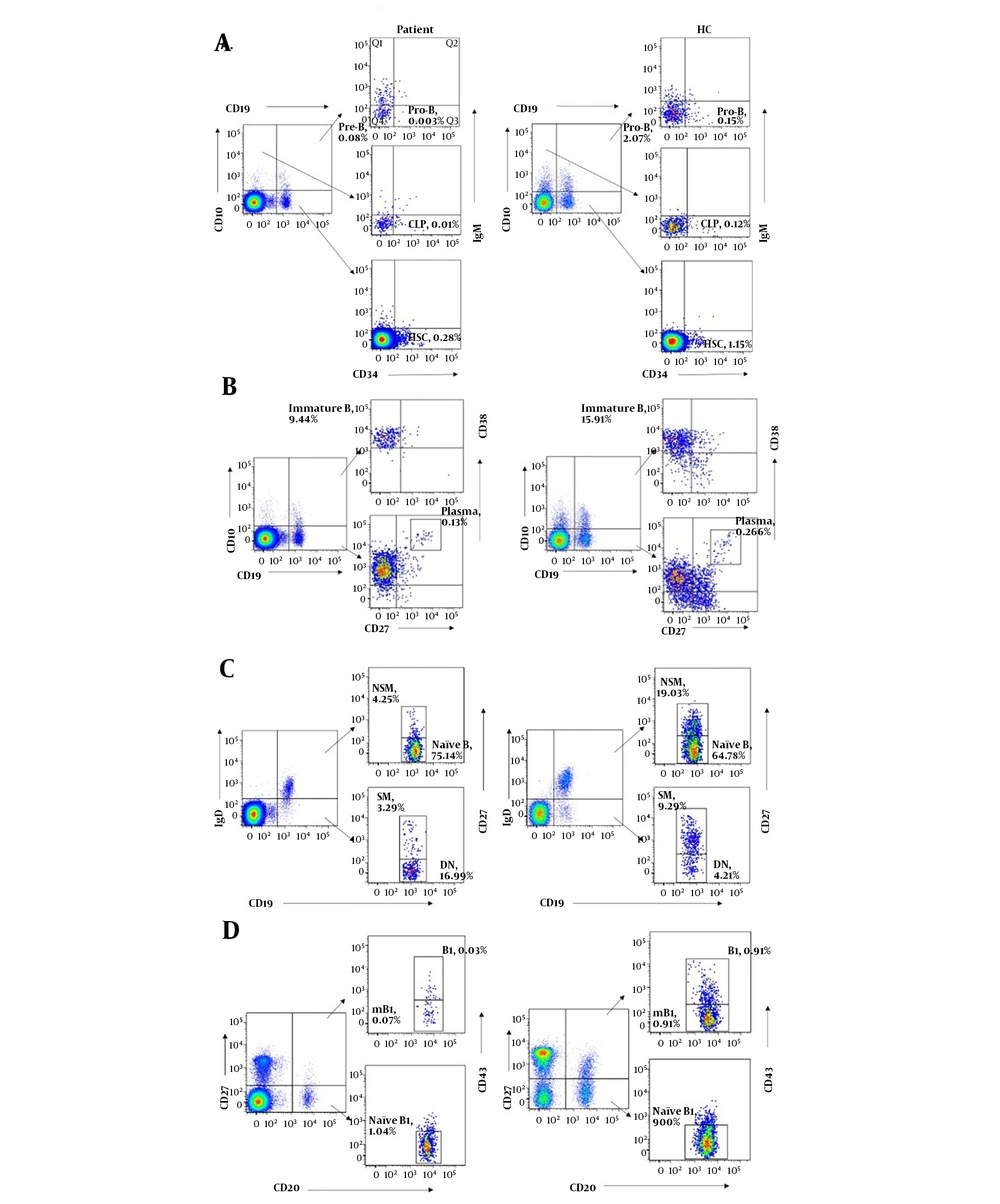
To understand the role of LRBA in B cell development, B cell immunophenotyping was performed in this study. Bone marrow progenitor B cell subsets in peripheral blood could be divided into HSC, CLP, Pro-B and Pre-B cells (13). Compared with age-matched healthy controls, all of the bone marrow progenitor B cell subsets (% of lymphocytes) were dramatically decreased in the patient. Besides, immature B cells were also decreased in the patient (Figure 3A and B and Table 2).
B cell immunophenotyping of the patient with LRBA deficiency and an age matched healthy controls. A, bone marrow progenitor B cell subsets (HSC, CLP, pro-B and Pre-B cells); B, B cell subsets in the antigen-dependent development period (immature B, mB and plasma cells); C, memory B cell subsets (naïve B, NSM, SM, DN); D, B1 subsets (naïve B1, B1 and mB1 cells). HSC, hematopoietic stem cell; CLP, common lymphoid progenitor; Pro-B, progenitor B cells; Pre-B, precursor B cells; NSM, non-switched memory B cells; SM, switched memory B cells; DN, double negative B cells; mB1, memory B1 cells.
| Age-Matched Healthy Controls | Patient in This Study | |
|---|---|---|
| Bone marrow progenitor B cell subsets, % of lymphocytes | ||
| HSC | 1.53 (1.15 - 1.96) | 0.28↓ |
| CLP | 0.15 (0.10 - 0.28) | 0.01↓ |
| Pro-B cells | 0.16 (0.13 - 0.19) | 0.00↓ |
| Pre-B cells | 2.01 (1.17 - 2.76) | 0.08↓ |
| B cell subsets, % of CD19+ cells | ||
| Immature B cells | 19.26 (11.61 - 30.72) | 9.44↓ |
| Naïve B cells | 65.44 (56.72 - 74.61) | 75.14↑ |
| Memory B cell | 26.6 (18.65 - 32.58) | 7.54↓ |
| Non-switched memory B cells | 13.4 (9.48 - 19.03) | 4.25↓ |
| Switched memory B cells | 13.17 (7.76 - 19.86) | 3.29↓ |
| Double negative B cells | 7.03 (4.21 - 12.84) | 16.99↑ |
| IgE+ B cells | 0.556 (0.24 - 0.85) | 9.63↑ |
| Plasma cells, % of lymphocytes | 0.39 (0.16 - 0.91) | 0.13↓ |
| B1 cell subsets, % of lymphocytes | ||
| Naïve B1 cells | 7.17 (5.05 - 9.00) | 1.04↓ |
| B1 cells | 0.8 (0.46 - 1.4) | 0.03↓ |
| Memory B1 cells | 2.22 (1.61 - 3.64) | 0.07↓ |
Percentages of B-Cell Subsets in the Patient and Age-Matched Healthy Controls (N = 5)
Memory B cell subsets could be sub-grouped for their expression of CD27 and IgD on their surface. CD27+IgD+ and CD27+IgD- B cells represented non-switched memory B cells and switched memory B cells, respectively (14). Compared with age-matched healthy controls, both non-switched and switched memory B cells were significantly decreased in the patient, while his naïve B cells (CD27-IgD+) and double negative B cells (CD27- IgD-) were increased, suggesting that memory B cells development was defective in the patient. Plasma cells were also slightly decreased in the patient. However, IgE+ B cells were greatly increased in the patient compared to the healthy controls (Figure 3B and C and Table 2). In addition, all of the B1 cell subsets (naïve B1, B1 and memory B1 cells) were found to be decreased in the patient, which indicated that the B1 cell development was also defective in the patient (Figure 3D and Table 2).
5. Discussion
LRBA deficiency was reported to have a broad and variable clinical phenotype, mainly including hypogammaglobulinemia, recurrent infection, autoimmune disorders, organomegaly and chronic diarrhea. Other clinical features included allergy and asthma, growth retardation, neurologic disease, and so on (2, 3, 5, 8). In a recent review including 32 LRBA deficient patients worldwide, 61% patients were reported to suffer from chronic diarrhea, autoimmune diseases, organomegaly and respiratory infections, and 58% patients from hypogammaglobulinemia (2). A higher frequency of these clinical manifestations was reported in a large Iranian cohort with LRBA deficient patients, which might be due to a longer follow-up period (3). Interestingly it has been reported that affected siblings with a same genetic defect in LRBA gene had different clinical manifestations, which highlights its potential role of as yet an unidentified modifier gene (15). In fact, because of the diversity of the clinical symptoms, there is still no standard diagnosis for LRBA deficiency (10).
In this study, we reported, for the first time, a Chinese patient with LRBA deficiency. The patient suffered from recurrent respiratory tract infections, EBV-associated lymphoproliferative disease and systemic vasculitis. Recurrent respiratory tract infection was one of the most frequently seen clinical symptoms of LRBA deficiency (2, 10). In a large cohort study of LRBA-deficient patients, 71% (15/22) patients had recurrent infections, in which recurrent upper and lower respiratory tract infections accounted for 48% (10/22) and 41% (9/22), respectively (10). Another large cohort of LRBA deficiency reported that 76.5% infected individuals suffered from pneumonia (3). In this study, the patient suffered from persistent lung infections for the past eight years. Except for the LRBA deficiency, the defective neutrophil activation might also contribute to the recurrent infection in the patient. In fact, although defective neutrophil respiratory burst has not been reported in LRBA deficient patients, it has been reported in other CVID patients (16). The neutrophil abnormalities might lead to an increased risk for recurrent infections.
At the age of 4 years, the patient was diagnosed as EBV infection. His EBV-associated symptoms included intermittent fever, lymphadenopathy and hepatosplenomegaly, which have been reported in patients with LRBA deficiency (1, 17). However, EBV infection was not frequently observed in LRBA deficient patients. Only 1/17 and 2/31 LRBA deficient patients were reported to suffer from EBV infection in two large cohort studies, respectively (2, 3). In addition, the patient was reported to have autoimmune disease which presented as autoimmune vascular inflammation. Autoimmune disease, which is one of the most frequent clinical phenotypes in LRBA deficient patients, was reported to occur in 76.5% and 61% patient with LRBA deficiency previously (2, 3). The most common features included AIHA (autoimmune hemolytic anemia), ITP (immune thrombocytopenia), IBD (inflammatory bowel disease) and so on (4, 10). In addition, the patient also had clinical presentations which have been frequently reported in LRBA patients previously, such as granulomatous lymphadenitis, sinusitis and oral candidiasis. Azizi et al. (3) reported lymphadenopathy, sinusitis and oral candidiasis occurred in about 52.9%, 70.6% and 23.5% patients, respectively; while a lower frequency was reported in Alkhairy et al. (2).
LRBA deficiency was firstly described to associate with early-onset hypogammaglobulinemia (8). However, later reports showed that not all of the LRBA mutant patients presented with hypogammaglobulinemia (9, 10, 15). In this study, the patient did not have hypogammaglobulinemia because of his normal serum IgG level, which seems contradictory to his decreased B cell counts. However, we have noticed that, compared to the healthy age matched controls, the plasma cells (% of lymphocytes) were only slightly reduced in this patient, which might, at least partially, explain the normal serum IgG level in the patient. Besides, the patient was reported to have extremely high level of IgE. To our knowledge, this is the first report of an association of LRBA deficiency with elevating serum IgE level. Our further finding showed that, although the patient had greatly decreased B cells, his IgE+ B cells (% of lymphocytes) are significantly elevated compared to the age-matched healthy controls, which could partially explain the hyper serum IgE in this patient.
Because of the great variety of the clinical features of LRBA deficiency, it is difficult for the clinician to distinguish it from other PIDs. Therefore, genetic analysis becomes the only way to make a definite diagnosis of LRBA deficiency. With the widespread use of next generation sequencing technology (NGS), the genetic diagnosis of LRBA deficiency becomes more feasible. NGS has many advantages and greatly improves the ability to identify gene defects in affected individuals (18). The application of targeted gene capture followed by NGS has more advantages, including cost saving, higher sequencing accuracy and shorter turnover time (19). More importantly, the data is more feasible both for the bioinformatics and further clinical analysis. Thus, it provides a more robust and effective way for the rapid clinical genetic diagnosis regarding human diseases, especially monogenetic disease (20).
As many subtypes of CVIDs, LRBA deficiency has been reported to associate with defective B cell development, but the underling mechanism still needs to be determined (8, 10). Gamez-Diaz et al. reported that most LRBA-deficient patients had low B-cell subset counts, mainly including switched memory B cells (80%) and plasmablasts (92%) (10). A further study showed that except for switched memory B cells and plasmablasts, naïve B cells, transitional B cells and marginal zone B cells all decreased in a majority of infected individuals (3). Besides, T cell deficiency, especially defects in Treg cells, was also reported in a considerable LRBA deficient patients. Azizi et al reported that, although 94.1% LRBA deficient patients in their cohort had normal T-cell counts, as high as 60% had decreased Treg cells (3). The patient in this study was shown to have normal T cell subsets, but unfortunately, his Treg cell counts were not detected.
The B cell immunophenotyping revealed that the patient’s HSC, CLP, Pro-B, Pre-B and immature B were all dramatically decreased, which suggested that LRBA gene affected the B cell development as early as the bone marrow development stage. Although the early B cell progenitors in peripheral blood could not fully represent precursor B cells in bone marrow, it did provide an evidence to demonstrate that the LRBA plays a key role in the precursor B cells development.
Naïve B cells (CD27-IgD+) differentiate into memory B cells and plasma cells in the germinal centers, where the antigen-activated naïve B cells undergo somatic hypermutation and isotype switching (21). Memory B cells consist of many phenotypic and functional heterogeneous subpopulations, and they are classically divided on the surface expression of CD27 and IgD: CD27-IgD+ as mature naive B cells, CD27+IgD+ as non-switched memory B cells, CD27+IgD- as switched memory B cells and CD27-IgD- as double negative B cells (14). Non-switched memory B cells are independent subsets of memory B cells, which play an important role in secondary immune response by prompt synthesizing high-affinity IgM (22). Switched memory B cells are able to produce IgG, IgM and IgA, which play a vital role in the humoral immunity (23). The origin and function of double negative B cells are debatable. Wei et al considered it was distinct lineage of memory B cells or a progenitor of memory CD27+ cells (24). Colonna-Romano el al. hypothesized that double negative B cells are senescent memory B cells which have down-regulated CD27 (25). In this study, both non-switched memory and switched memory B cells in the patient were significantly decreased, while his naïve and double negative B cells were increased, which indicated a defective memory B cell development in the patient. Similarly, Lopez-Herrera et al. reported LRBA deficient patients had low switched memory B-cell cells (8). Gamez-Diaz et al. also observed decreased switched memory B-cells in most (80%) of the LRBA deficient patients (10). In fact, decreased CD27+ memory B cells have been reported in most CVID patients with non-LRBA deficiency (26). However, the underlying mechanism remains unclear.
B1 cells are a predominant cell population in body cavities. They produce natural antibody, which are known to participate in immune responses against bacteria, viruses and certain parasites. The patient in this study had greatly decreased B1 cell subsets, which indicated that LRBA deficiency played an important role in B1 cell development. Although defective B1 cell development has not been reported in LRBA deficiency, it has been reported in other CVID patients (27).
Taken together, we reported a patient with compound heterozygous mutations in LRBA gene in China, who presented with EBV infection, systemic vasculitis, recurrent infection and hyper serum IgE. Our results indicated the importance of LRBA in B cell development. Further studies are still needed for a better understanding of the biological functions of LRBA gene.