1. Background
Hypospadias (HS) has been defined as a condition in which the urethral meatus may be proximal to its normal glanural occurrence on the ventral surface of the penis, on the scrotum or the perineum. In humans HS is the second most common congenital anomaly with an incidence of 1 in 250 - 300 live male births and its pathogenesis is complex, multifactorial, and determined by genetic, hormonal and environmental causes (1). There is an increasing interest in the concept that maternal use of preparations containing folic acid and other water-soluble vitamins may influence the occurrence of this abnormality in offspring (2, 3). The presence of HS has severe consequences on both physical and psychological development and living situation (1, 4). Complications of corrective surgery in HS correlate with patients’ opinions on their voiding ability and sexual life as adults (5, 6).
Recently, Douglas et al. (7) reported a requirement for 6(R)-L-erythro-5,6,7,8-tetrahydrobiopterin (BH4) in embryonic development. BH4, a metabolite structurally related to riboflavin and folic acid by sharing the common pterin backbone, is synthesized de novo from guanosine triphosphate (GTP) through the catalysis of three enzymatic reactions by GTP cyclohydrolase (GTPCH, EC 3.5.4.16; rate-limiting step), 6-pyruvoytetrahydrobiopterin synthase, and sepiapterin reductase (8). GTPCH deficiency, due to loss of function mutations in the GCH1 gene, results in neurological disorders (7-9). Urine profiles of pterin compounds have the potential to become predictive biomarkers of bladder cancer (10). Moreover, Acevedo-Alavarez and colleagues (11) reported that mouse urothelial transcripts of Gch1 might play an important role in modulating voiding behavior changes.
BH4 is an important antioxidant and an essential cofactor for three isoforms of nitric oxide (NO) synthase and three aromatic amino acid hydroxylases, including phenylalanine hydroxylase (PAH, EC 1.14.16.1), as well as highly hydrophobic alkylglycerol monooxygenase (AGMO, also called glyceryl-ether monooxygenase, EC 1.14.16.5). AGMO is the only enzyme known to cleave the ether bond in alkylglycerol ether lipids. Alkylglycerols are a subclass of lipids, which are membrane constituents and are involved in many signaling pathways and in spermatogenesis (12).
Marrocco et al. (1) hypothesized in their recent review article that most patients develop HS because of interactions between environmental stimuli and polymorphic variants of genes. HS is associated with a high rate of extra urogenital anomalies, e.g., congenital heart defects, orofacial clefts and spina bifida (13-15), leading to the anticipation that there may be a partly shared genetic background for these congenital anomalies. Polymorphic variants of GCH1 are associated with the pathogenesis of spina bifida (16) and orofacial clefts (9). The involvement of the PAHgen in the etiology of orofacial clefts was reported in the Polish population (17). AGMO belongs to candidate genes for congenital heart anomalies in humans (18). Smith-Lemli-Opitz is an inherited autosomal recessive syndrome resulting from a defect in cholesterol synthesis. This syndrome is characterized by mental retardation and associated multiple anomalies including hypospadias (19). It is noteworthy that ether lipids deficiency impairs intracellular cholesterol distribution and homeostasis (12, 20). An association between diabetes and HS was suggested, although this issue is still debated (21). Of particular interest is the finding that polymorphic variant rs2191349, which lies between the AGMO and the DGKB gene, is associated with reduced glucose-stimulated insulin response at a genome-wide significance level (22, 23).
2. Objectives
We examined whether polymorphic variants in GCH1, PAH and AGMO/DGKB genes were associated with hypospadias risk in the Polish population.
3. Methods
3.1. Study Population
Cases were boys treated for HS at the Department of Paediatric Surgery of the Warsaw’s Institute of Mother and Child. The medical records were reviewed to obtain clinical data on HS phenotype and coexisting congenital malformations. Cases with a known cause of HS were excluded. To ensure independent analyses, only one case or control per family was included in the study by excluding youngest brothers or one of the brothers at random for same-sex twins. The study was approved by the local Ethic Committee. Oral and written consent was obtained from the legal guardians of all of the participants.
Previous research showed the familial occurrence of hypospadias for anterior and middle forms of that malformation but not for posterior types, pointing toward genetic risk factors being important (3). Because of this observations, we included only isolated anterior and middle cases to the current study. A total of 166 boys (13 months to 10 years old) presenting with isolated hypospadias and 285 unrelated boys without congenital malformations, were recruited. The ancestry contribution was estimated at 100% of Caucasians of the Polish descent in the case and the control group. DNA was isolated from peripheral blood lymphocytes by a salt-induced extraction procedure.
3.2. Single Nucleotide Polymorphism (SNP) Selection and Genotyping
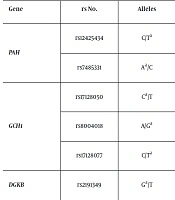
Five SNPs in GCH1 and PAH, previously detected to be in association with orofacial clefts risk in the Polish population (9, 17), as well as rs2191349 of the AGMO-DGKB loci were evaluated in this study (Table 1). The genotyping was carried out by high-resolution melting curve analysis (HRM) on the Light Cycler 480 system (Table 2). For quality control, approximately 10% of the randomly chosen samples were re-genotyped. Samples that failed genotyping were not repeated and were removed from statistical calculations.
| Gene | rs No. | Locationa | Allelesb | SNP Functionc | MAFd |
|---|---|---|---|---|---|
| PAH | rs12425434 | chr12:103240067 | C/Tb (FWD) | intron | 0.27 |
| rs7485331 | chr12:103312619 | Ab/C (FWD) | N/A (upstream) | 0.29 | |
| GCH1 | rs17128050 | chr14:55343879 | Cb/T (FWD) | intron | 0.10 |
| rs8004018 | chr14:55350696 | A/Gb (FWD) | intron | 0.10 | |
| rs17128077 | chr14:55386034 | C/Tb (FWD) | N/A (upstream) | 0.11 | |
| DGKB | rs2191349 | chr7:15064309 | Gb/T (FWD) | N/A (upstream) | 0.44 |
Abbreviation: FWD, forward.
aNCBI build 37/hg19.
bDenotes the minor allele (based on whole sample).
cAccording to the Single Nucleotide Polymorphism database (dbSNP).
dMAF, minor allele frequency calculated from the control samples.
| Gene | rs No. | Allelesa | Primers for PCR Amplification (5’ - 3’) | Annealing Temp. (°C) | PCR Product Length (bp) | Melt. Temp. Range (°C) |
|---|---|---|---|---|---|---|
| PAH | rs12425434 | C/Ta | F: ATTTGCACTCATTGGCAGTCC | 60.6 | 68 | 68 - 80 |
| R: ATTGCCTGTCCTGGAAGTTGA | ||||||
| rs7485331 | Aa/C | F: TTCCCATAGTAAGTTGGAAGC | 60.6 | 146 | 76 - 86 | |
| R: TGAGGCTGAGGAATACAACA | ||||||
| GCH1 | rs17128050 | Ca/T | F: GCTCCAACATATCTAAAAGCTACCA | 55.0 | 103 | 72 - 87 |
| R: GGGTTACCTTCTTGCTGCTG | ||||||
| rs8004018 | A/Ga | F: TTAAAAATTTGTGAGGAC | 53.0 | 109 | 75 - 90 | |
| R: ATTGATTTCTAATGAGTTGG | ||||||
| rs17128077 | C/Ta | F: ATGGAATCTAAGGCCATGTTCAGC | 58.0 | 115 | 75 - 90 | |
| R: AGACCAGCCTGGGACACATGA | ||||||
| DGKB | rs2191349 | Ga/T | F: AGGCCTTAACTTTGGCTGGA | 55 | 81 | 75 - 90 |
| R: AGACCCCACCGCTAGATGTT |
aDenotes the minor allele (based on whole sample).
3.3. Statistical Methods
For each SNP, the Hardy-Weinberg (HW) equilibrium was evaluated in both patients and controls by the chi-square (χ2) test. Statistically significant deviation from HW expectations was interpreted as P value < 0.05. The differences in allele frequencies between cases and controls were determined using standard χ2 test. The strength of association was estimated by odds ratio (OR) and corresponding 95% confidence intervals (95%CIs). The dominant and recessive models were tested. To account for multiple comparisons the strict Bonferroni correction was applied. The P values below 0.0083 (0.05/6 SNPs) were considered as statistically significant.
The haplotype-based association analysis using a sliding window approach was conducted using Haploview V. 4.0 software. Significant P values were corrected using the 10,000-fold permutation test.
Higher-order gene-gene interactions among all tested single nucleotide polymorphisms were evaluated using the non-parametric and genetic model-free multifactor dimensionality reduction (MDR) approach (MDR version 3.0.2).
4. Results
None of the tested nucleotide variants showed evidence for deviation from Hardy-Weinberg equilibrium in either cases or controls. The minor allele frequency (MAF) for all SNPs was ≥ 10% (MAF calculated from the controls, Table 1). Genotype counts, OR and 95% CI calculations for the all tested SNPs are reported in Table 3. Overall, there was no evidence for either allelic or genotypic association with the risk of being born with HS. Haplotype analysis of nucleotide variants in the GCH1 and PAH locus revealed no haplotypes associated with the risk of HS (Table 4). Results of exhaustive MDR analysis evaluating combinations of all tested SNPs are summarized in Table 5. The combinations did not reach statistical significance in predicting susceptibility to HS.
| Gene | rs No. | Allelesa | Genotype Distributionb, MAF | Ptrend | Pallelic | Pgeno | Dominant Modelc | Recessive Modeld | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | OR (95% CI) | P Value | OR (95% CI) | P Value | ||||||
| PAH | rs12425434 | C/Ta | 16 / 59 / 91 | 21 / 111 / 147 | 0.9975 | 0.9975 | 0.5664 | 0.918 (0.624 - 1.350) | 0.6629 | 1.310 (0.663 - 5.589) | 0.4352 |
| 0.27 | 0.27 | ||||||||||
| rs7485331 | Aa/C | 13 / 63 / 88 | 22 / 120 / 138 | 0.4905 | 0.4930 | 0.6426 | 0.839 (0.570 - 1.235) | 0.3737 | 1.010 (0.494 - 2.063) | 0.9790 | |
| 0.27 | 0.29 | ||||||||||
| GCH1 | rs17128050 | Ca/T | 3 / 29 / 134 | 2 / 54 / 224 | 0.9305 | 0.9303 | 0.5216 | 0.955 (0.589 - 1.550) | 0.8529 | 2.558 (0.423 - 15.478) | 0.3659e |
| 0.11 | 0.10 | ||||||||||
| rs8004018 | A/Ga | 3 / 29 / 131 | 2 / 54 / 225 | 0.8456 | 0.8453 | 0.5272 | 0.982 (0.604 - 1.594) | 0.9397 | 2.616 (0.432 - 15.826) | 0.3616e | |
| 0.11 | 0.10 | ||||||||||
| rs17128077 | C/Ta | 4 / 36 / 126 | 5 / 52 / 223 | 0.3440 | 0.3304 | 0.6377 | 1.242 (0.784 - 1.967) | 0.3548 | 1.358 (0.359 - 5.131) | 0.7324e | |
| 0.13 | 0.11 | ||||||||||
| AGMO | rs2191349 | Ga/T | 30 / 85 / 50 | 66 / 118 / 97 | 0.8790 | 0.8744 | 0.1359 | 1.213 (0.802 - 1.833) | 0.3604 | 0.724 (0.447 - 1.173) | 0.1881 |
| 0.44 | 0.44 | ||||||||||
Abbreviation: MAF, minor allele frequency.
aDenotes the minor allele (in all cases the minor allele is the risk allele).
bThe order of genotypes: dd/Dd/DD (d is the minor allele).
cDominant model: dd + Dd vs. DD (d is the minor allele).
dRecessive model: dd vs. Dd + DD (d is the minor allele).
eFisher exact test.
| Gene | Polymorphisms | Haplotypes | Frequency | Case, Control Ratios | Chi-Square | P Value | Pcorr Valuea |
|---|---|---|---|---|---|---|---|
| PAH | rs12425434-rs7485331 | CC | 0.445 | 0.456, 0.438 | 0.256 | 0.613 | 0.858 |
| CA | 0.281 | 0.270, 0.288 | 0.320 | 0.572 | 0.837 | ||
| TC | 0.270 | 0.273, 0.269 | 0.015 | 0.901 | 0.996 | ||
| GCH1 | rs17128050-rs8004018 | TA | 0.896 | 0.895, 0.896 | 0.008 | 0.930 | 1.000 |
| CG | 0.104 | 0.105, 0.104 | 0.008 | 0.930 | 1.000 | ||
| rs8004018-rs17128077 | AC | 0.861 | 0.845, 0.871 | 1.121 | 0.290 | 0.674 | |
| GT | 0.085 | 0.086, 0.085 | 0.002 | 0.966 | 1.000 | ||
| AT | 0.033 | 0.047, 0.026 | 2.840 | 0.092 | 0.247 | ||
| GC | 0.020 | 0.022, 0.019 | 0.134 | 0.715 | 0.976 | ||
| rs17128050-rs8004018-rs17128077 | TAC | 0.861 | 0.846, 0.871 | 1.092 | 0.296 | 0.722 | |
| CGT | 0.085 | 0.084, 0.085 | 0.005 | 0.945 | 1.000 | ||
| TAT | 0.034 | 0.049, 0.026 | 3.375 | 0.066 | 0.197 | ||
| CGC | 0.020 | 0.022, 0.019 | 0.110 | 0.740 | 0.982 |
aP value calculated using permutation test and a total of 1,000 permutations.
| Genes and rs Numbers | Testing Balanced Accuracy | Cross Validation Consistency | P Valuea |
|---|---|---|---|
| PAH-rs7485331, DGKB-rs2191349 | 0.4797 | 7 / 10 | 0.943 |
| PAH-rs12425434, PAH-rs7485331, DGKB-rs2191349 | 0.4988 | 8 / 10 | 0.846 |
| PAH-rs12425434, PAH-rs7485331, GCH1-rs17128077, DGKB-rs2191349 | 0.5299 | 7 / 10 | 0.535 |
aSignificance of accuracy, empirical p value based on 1,000 permutations.
5. Discussion
The network-oriented picture of HS risk factors is complex and likely largely incomplete (1). Non-functional mutations of the GCH1, resulting in complete loss of BH4 synthesis, have not been described, suggesting that complete loss of GTPCH activity, as well as BH4-dependent enzymes activity, is likely to be embryonically lethal in humans (7). Our study addressed two questions. First, are SNPs of GCH1 and PAH that were nominally significant upon testing for association with birth defects known for frequent occurrence with hypospadias also associated with isolated hypospadias? Second, is the intensively studied SNP associated with abnormal glucose homeostasis linked to hypospadias susceptibility? To our knowledge, the present report is the first association study evaluating nucleotide variants of the BH4 pathway genes as possible risk factors for HS.
A study conducted by Lupo and colleagues (16) suggested a link between haplotypes of GCH1 gene and abnormal neural tube closure in the USA. In our previous study, we found a significant association between the GCH1 rs17128077, rs8004018 and rs17128050 variants with an increased risk for cleft lip with or without cleft palate (9). The correlation with single polymorphic variants was confirmed at the haplotype level (9). The investigated variants of the GCH1 were not found to be involved in the pathogenesis of HS.
The PAH rs7485331 and rs12425434 were reported to influence susceptibility to isolated orofacial clefts (17). Mutations in the PAH gene cause phenylketonuria, the most common inborn error of amino acids metabolism in Middle East Asians and Europeans. In the recent description of 30 patients with phenylketonuria and co-existent disorders from 13 treatment centers from Europe and Turkey, one boy suffered from hypospadias (24). In untreated or non-optimally treated pregnancies of phenylketonurics, HS in offspring as a less frequent sign of maternal phenylketonuria syndrome was described (25). However, in our study statistical analysis revealed no significant association between common nucleotide variants of PAH and the risk of HS.
The sequence of AGMO shows no significant similarity with the other known BH4-dependent enzymes but contains the typical fatty acid hydroxylase protein motive signature (26). The SNP rs2191349 of the AGMO-DGKB loci was not associated with abnormal penile development in the present study. Recent studies revealed no association between this polymorphic variant and cognitive function in children (27) and chronic kidney disease in patients with type 2 diabetes (28).
It is worth noting that negative results of single-marker analysis for GCH1 and PAH were confirmed by haplotype evaluation. Variation in populations is inherently structured into haplotypes (29). Analysis of epistasis is useful in determining the true contribution of genetic factors to birth defect susceptibility (30). All of the genes we examined have not been studied previously for their contribution to HS. In contrast to preliminary expectations, we did not identify the significant interactive effect of polymorphic variants of BH4 pathway genes on HS risk.
An important strength of presented study is the use of data from individuals recruited from the homogeneous population. The major limitation is the sample size, which did not allow us to detect modest interactions and associations. We must also note that the number of selected SNPs does not cover the GCH1 and PAH genes fully and extensively. Additional extensions and modifications of the MDR approach will be needed to fully address the complexity of genetic association data.
In conclusion, the presented results did not support any association between polymorphic variants of GCH1, PAH and AGMO-DGKB loci, and risk of being born with hypospadias in the Polish population.