1. Background
Acute lymphoblastic leukemia (ALL) is the most conventional hematologic malignancy in children, with an incidence maximum at 2 to 5 years of age (1). Aberrant genetic or epigenetic changes can lead to an interruption of differentiation and uncontrolled expansions of immature thymocytes in T-cell development (2). One of the key oncogenic signals in the initial T-cell development and T-ALL is Notch, which is efficiently active in more than 50% of T-ALLs (3). Notch signaling pathway regulation happens by its intra membrane proteolysis. Released intracellular fragment participates directly in transcriptional regulation of nuclear target genes.
Regulatory errors in Notch signaling are linked to numerous human disorders, such as cardiovascular disorders and malignancy (4). Regulated Notch function is crucial for the proliferation of T-cells, atypical Notch signaling leads to leukemia (5). Recently, an increasing number of analyses displayed that the miRNA expression pattern in ALL plays a significant role in the Notch1 regulatory route during the progress of leukemia (6, 7).
Mir-200 family members, located on human chromosomes 1 and 12, include five microRNAs: miR-200a, 200b, 200c and miR-141, 429 (8) and are known as regulators of the epithelial phenotype through suppression of ZEB1 and ZEB2 mRNA translation (9). This miRNA family has also been shown to have pleiotropic effects, including regulation of stem cell factors and features, indicative of their importance for tissue homeostasis (10). The 5- Cytosine connected to guanine by phosphodiester bond (5-CpG islands) hypermethylation-correlated silencing of both miR-200 loci is noticed in transformed cells with mesenchymal characteristics, consisting of low levels of ZEB1/ZEB2 and high E-cadherin expression, mesenchymal-epithelial transitions (MET) phenotype in tumor development (11). The miR-200 cluster, which targets Jagged1, has been described as a key regulator of the Notch pathway in cancer (8). In addition, miR-200 and miR-205 repression leads to activation of Notch signaling (12). In the hematopoietic system, Notch directly regulates the differentiation and maturation of normal T lymphocytes, the discrimination between the CD4 and CD8 lineages, and the expression of TCRα/β receptors (13, 14). Wang et al. showed that transfection of miR-200b in pancreatic cell line has reduced the levels of Jagged-1/2 and those of their target genes Hes-1, Hey-2 and Bcl-2 leading to cell growth inhibition (15).
The miR-34 cluster stimulates via the tumor suppressor p53 and is recognized to prevent epithelial-mesenchymal transition (EMT), and so, doubtless suppresses the initial stages of metastasis (16). Repression of miR-34a/b/c affects up-regulation of SNAIL (Snail family transcriptional repressor 1) with the show of EMT indicators and correlated features and increased migration or invasion. MiR-34a also represses SLUG (Snail family transcriptional repressor 2) and ZEB1 (17). MiR-34 cluster is placed in 11q position. Actually, down-regulation of miR-34a in CLL has been associated with p53 silence, reduced DNA damage response, and apoptosis resistor (18). MiR-34a is tumor suppressor miRNA with capability to adjust the expression of several targets known in tumorigenesis and cancer progress such as MET, MYC, CDK4/6, Notch1, BCL2, CD44 and various other molecules (19). Di Martino et al. have also indicated the possible miR-34a action in down-regulating both Bcl-2 and Notch1 and activation of apoptosis both in vitro and in mouse models (20).
Recent studies have displayed the deregulation of miRNA expression and contribution of miRNAs to the multistep processes of carcinogenesis such as oncogenes or tumor suppressor genes (TSGs) in human tumor (21). Also have extensively demonstrated that hypermethylation of promoters in ALL is a frequent mechanism of gene silencing and is associated with the prognosis and the response to treatment (22).
2. Objectives
In present study, we have investigated the relationship between the hypermethylation of promoter of suppressor gene of mir-200b and miR-34a with the Notch signal pathway in acute lymphoblastic leukemia patients.
3. Methods
3.1. Patients and Clinical Samples
We collected blood samples from patients diagnosed with ALL by observing ethical standards and obtaining consent from parents. Samples were obtained from patients referred to Mahak Hospital in Tehran, Iran, from 2015 to December 2017. University Ethical Committee approved this study (approval number: IR.IAU.Z.REC.1397.060/061). Participants’ clinical data included age, sex, BMI, stage of treatment, cancer type, white blood cells, platelets count and hemoglobin level.
Thirty blood samples were collected from patients with ALL with an average age of 5.43 years (range, 1 to 16 years). Patients were divided into two groups before treatment (new cases) and during treatment (under the control), 6 of these were repeated in the new cases and those under the control phase. Normal samples (n = 30) were collected from healthy participants with an average age of 6.163 years (range, 1 month to 13 years). These participants did not have any history of leukemia or clinical symptoms and were not related with the patients.
3.2. DNA Extraction and Methylation Specific PCR (MSP)
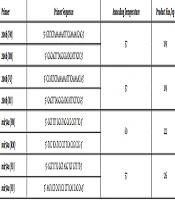
DNA was extracted from peripheral blood lymphocytes of the patients and normal controls using Cinaapure DNA (Cat.No.PR881612). After sodium bisulfite treatment of DNA, DNA methylation patterns in the mir-200b and mir-34a promoter region were determined using methylation-specific PCR (MSP). During bisulfite treatment, the unmethylated cytosines are modified to thymine while methylated cytosines remain unchanged. MSP analyses are capable of distinguishing homozygous or heterozygous methylation status in the samples. This method is based on the use of two primers to separate methylated from unmethylated DNA in bisulfite-modified DNA. Bisulfite modification of genomic DNA was done using EpiTect Plus DNA Bisulfite Kit (Cat No. /ID: 59124, QIAGEN Inc), according to manufacturer’s constructions. Primer sequences for selected microRNAs are shown in Table 1.
| Primer | Primer Sequence | Annealing Temperature | Product Size, bp |
|---|---|---|---|
| 200b (FM) | 5’- GTCTCTAAAAAAATTTCGAAAACGAC-3’ | 57 | 189 |
| 200b (RM) | 5’- GGGAGTTTAGGGGATATATTTGTC-3’ | ||
| 200b (FU) | 5’- CCATCTCTAAAAAAATTTCAAAAACA-3’ | 57 | 189 |
| 200b (RU) | 5’- GAGTTTAGGGGATATATTTGTTGG-3’ | ||
| mir34a (FM) | 5’- GGT TTT GGG TAG GCG CGT TTC-3’ | 60 | 122 |
| mir34a (RM) | 5’- TCC TCA TCC CCT TCA CCG CCG -3’ | ||
| mir34a (FU) | 5’- GGTT TTG GGT AGG TGT GTT TT-3’ | 57 | 126 |
| mir34a (FU) | 5’- AAT CCT CAT CCC CTT CAC CAC CA-3’ |
Summary of Primer Sequences and Annealing Temperatures for 200b Promoter Regions and PCR Product Sizes for MSP
Methylation specific polymerase chain reaction (MSP) of mir200b and mir-34a genes were performed with two primers designed for methylated and unmethylated regions. PCR reaction mix (20 µL) included 1X taq premix 10 µL, upstream primer (10 mol/µL) 1 µL, downstream primer (10 mol/µL) 1 µL, template DNA (100 ngr) 2 µL, sterilized DW 6 µL. Thermal cycling conditions were as follows: an initial denaturation step at 95°C for 5 minutes, followed by 35 cycles of 95°C denaturation for 45 seconds, annealing for 45 seconds (primer specific temperatures are listed in Table 1) and extensions at 72°C for 45 seconds, with a final extension at 72°C for 5 minutes. The PCR amplification products were electrophoresed on 2% agarose gel and visualized using the Image Analyzer, after staining with SYBR Safe DNA gel stain. If only unmethylated bands were seen during electrophoresis of MSP products, the sample was recorded as unmethylated; if methylated bands were seen, the sample was recorded as methylated; if unmethylated and methylated bands were observed, the sample was recorded as hemimethylated.
3.3. Statistical Analysis
All statistical analyses were performed with the Statistical Package for Social Sciences (SPSS, version 20). The differences between gene methylation status and Clinical Characteristics were evaluated using Pearson’s chi-square test. The association between hypermethylation of the genes and risk of ALL was estimated by calculating odds ratios (ORs) and 95% confidence intervals (CI) using the chi-square test and Fisher’s exact test. The association was considered as statistically significant if P < 0.05.
4. Results
Clinicopathological characteristics of patients (n = 30) were studied. Pathological diagnosis revealed that 18 were Pre-B and 1 Pro-B and 11 T-Cell according to WHO classification. Family history of cancer was reported in 9 of 30 cases. A total of 50% of patients were ≤ 6 years old, while 50% of patients were > 6 years old and only six of them were girls. Fatigue, weakness, fever, bone pain, hemoglobin level lower than 10 g/dL, platelet count (PLT) less than 100,000/µL were common symptoms of patients. Also white blood cell counts of patients were higher or lower than normal.
4.1. Methylation Analysis
The methylation status of the promoter region determined in 60 samples included 30 samples of healthy controls and 30 patients. Six patients were studied in the new case group and also during treatment. MSP data revealed that mir-200b and mir-34a genes promoter were methylated in 50% (15/30) and 30% (9/30), hemimethylated in 50% (15/30) and 56.6% (17/30) respectively, whereas un-methylation was observed in 0.0% and 13.4% (4/30) in patients’ samples. In comparison, control samples were methylated in 6.6% (2/30) and 10% (3/30), hemimethylated in 73.3% (22/30) and 30% (9/30), respectively; also un-methylated in 20% (6/30) and 60% (18/30). Our data showed that methylation of mir200b and mir-34a genes promoter significantly was associated with acute lymphoblastic leukemia (P < 0.0001 and P < 0.004, respectively) (Table 2).
| mir-200b | mir-34a | |||||
|---|---|---|---|---|---|---|
| Healthy, N = 30 | Patient, N = 24 | P Value | Healthy, N = 30 | Patient, N = 24 | P Value | |
| Methylated | 2 (3.7) | 13 (24.1) | 0.000 | 3 (10) | 6 (25) | 0.004 |
| Hemimethylated | 22 (40.7) | 11 (20.3) | 9 (30) | 14 (58.33) | ||
| Unmethylated | 6 (11.1) | 0 (0.0) | 18 (60) | 4 (16.66) | ||
Methylation Status of mir200b and mir34a in Patients and Healthy Controlsa
We evaluated the relationships of aberrant hypermethylation with clinicopathological factors (Table 3). Our analysis showed that hypermethylation of mir-200b gene was associated with family history (P = 0.003) and platelets (P = 0.01) in ALL. Also there was a significant association of mir-34a gene methylation with patient status (P = 0.003) and Hb (P = 0.001), while no association was observed between age, WBC, platelets, blast lineage and the hypermethylation of any gene (Table 3).
| Feature | mir-200b | mir-34a | ||||||
|---|---|---|---|---|---|---|---|---|
| Methylated | Hemimethylated | Unmethylated | P Value | Methylated | Hemimethylated | Unmethylated | P value | |
| Age, y | 0.7 | 0.48 | ||||||
| < 1 (1) | 1 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (100) | 0 (0.0) | ||
| 1 - 3 (6) | 4 (66.7) | 2 (33.3) | 0 (0.0) | 3 (50) | 2 (33.3) | 1 (16.7) | ||
| 4 - 10 (16) | 8 (50) | 8 (50) | 0 (0.0) | 3 (18.7) | 10 (62.5) | 3 (18.7) | ||
| > 10 (1) | 0 (0) | 1 (100) | 0 (0.0) | 0 (0.0) | 1 (100) | 0 (0.0) | ||
| WBC × 109/L | 1 | 0.4 | ||||||
| < Normal (8) | 4 (50) | 4 (50) | 0 (0.0) | 3 (37.5) | 5 (62.5) | 0 (0) | ||
| Normal (6) | 3 (50) | 3 (50) | 0 (0.0) | 2 (33.33) | 2 (33.33) | 2 (33.33) | ||
| > Normal (10) | 6 (63.6) | 4 (36.4) | 0 (0.0) | 4 (40) | 5 (50) | 1 (10) | ||
| Patients’ status | 0.7 | 0.003 | ||||||
| New case (16) | 8 (50) | 8 (50) | 0 (0) | 9 (56.2) | 7 (43.8) | 0 (0) | ||
| Treated (8) | 5 (62.5) | 3 (37.5) | 0 (0) | 0 (0) | 5(62.5) | 3 (37.5) | ||
| Stage of cancer | 0.3 | 0.7 | ||||||
| Relapse (4) | 3 (75) | 1 (25) | 0 (0.0) | 0 (0.0) | 3 (75) | 1 (25) | ||
| Induction (1) | 1 (100) | 0 (0.0) | 0 (0.0) | 1 (100) | 0 (0.0) | 0 (0.0) | ||
| Consolidationn (7) | 3 (42.9) | 4 (57.1) | 0 (0.0) | 0 (0.0) | 5 (71.4) | 2 (28.6) | ||
| Maintenance (2) | 0 (0.0) | 2 (100) | 0 (0.0) | 0 (0.0) | 1 (50) | 1 (50) | ||
| Platelet (24) | 0.01 | 1 | ||||||
| Abnormal (24) | 6 (25) | 1 (4.2) | 0 (0.0) | 3 (12.5) | 3 (12.5) | 1 (4.1) | ||
| Normal (5) | 0 (0.0) | 5 (20.8) | 0 (0.0) | 2 (8.4) | 3 (12.5) | 0 (0.0) | ||
| Dangerous (12) | 7 (29.2) | 5 (20.8) | 0 (0.0) | 4(33.3) | 6 (50) | 2(16.7) | ||
| Hemoglobin (24) | 0.8 | 0.016 | ||||||
| Abnormal (18) | 10 (55.6) | 8 (44.4) | 0 (0.0) | 8 (0.0) | 10 (0.0) | 0 (0.0) | ||
| Normal (5) | 3 (60) | 2 (40) | 0 (0.0) | 1 (20.0) | 2 (40.0) | 2 (40.0) | ||
| Dangerous (1) | 0 (0.0) | 1 (100) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (100) | ||
| Blast lineage (24) | 0.7 | 0.7 | ||||||
| Pre-B (15) | 8 (53.3) | 7 (46.7) | 0 (0.0) | 7 (46.7) | 6 (40) | 2 (13.3) | ||
| Pro-B (1) | 0 (0.0) | 1 (100.0) | 0 (0.0) | 0 (0.0) | 1 (100.0) | 0 (0.0) | ||
| T cell (8) | 5 (62.5) | 3 (37.5) | 0 (0.0) | 2 (25) | 5 (62.5) | 1 (12.5) | ||
| Family history history (24) | 0.03 | 0.7 | ||||||
| No (16) | 6 (37.5) | 10 (62.5) | 5 (31.2) | 10 (62.5) | 1 (6.3) | |||
| Yes (8) | 7 (87.5) | 1 (12.5) | 4 (50) | 2 (25) | 2 (25) | |||
In this study the methylation pattern of the promoter region for mir200b and mir-34a genes in 14 of 30 patients with leukemia who were in the stages of consolidation (n = 7), induction (n = 1), maintenance (n = 2) and relapse (n = 4) of treatment phases showed that the stage of treatment did not have a significant relationship with leukemia (P = 0.3, P = 1).
Six of the 30 patients were studied in the new cases group and also during treatment. The comparison of methylation pattern before and after treatment showed that 2 of 6 patients who were treated with vincristine (1.5 mg/m2/IV), mercaptopurine (50 mg/m2), and methotrexate (20 mg/m2) in the maintenance stage had been changed from methylated to hemimethylated in the mir-200b gene and from methylated to hemimethylated and unmethylated in the mir-34a gene. Also 4 of 6 patients who were treated with L-asparaginase (5000 u/m2/infusion) and vincristine (1.5 mg/m2/IV) in the consolidation stage showed that methylation pattern in 2 of these patients did not change in the mir-200b gene but in one patient it changed from methylated to hemimethylated and in another patient from hemimethylated to methylated. The pattern of methylation of mir-34a gene promoter in 3 of 4 patients did not change and in one patient it had changed from methylated to hemimethylated in the consolidation stage. The type of medication and their prescribing time at each stage are listed in Table 4.
| Medication | Stage of the Cancer |
|---|---|
| L-Asparaginase | Consolidation |
| Vincristine | Consolidation and maintenance |
| Cytarabine | Induction |
| Mercaptopurine | Induction and maintenance |
| Methotrexate | Induction and maintenance |
| Cyclophosphamide | Induction |
| 5-fluorouracil/leucovorin | Induction |
Drugs and Their Prescribing Time at Each Stage of Cancer
5. Discussion
According to literature, DNA methylation tends to happen in the promoter region of the genes and is one of the most studied epigenetic anomalies in the oncogenesis (23, 24). In recent years miRNAs have become definitely recognized as key molecular components of the cell in both normal and pathologic conditions (25). MiRNA genes are connected to CpG islands, which terminate to the great mechanism that can lead to abnormal epigenetic alteration (26). Changes in expression of miRNAs has been contributed to leukemogenesis and seems to have an effect on regulatory pathways of growth in ALL. Several specific miRNAs associated with pediatric ALL consist of miRNA (miR) miR-34, miR-128, miR-142, and miR-181, all stated to be more expressed (27, 28).
Our results have indicated that promoter methylation of mir-200b and mir-34a genes are related with ALL. Also this study indicates the role of promoter hypermethylation status of these two genes in carcinogenesis and molecular predictor of cancer progress. Wiklund et al. in 2011 found that in muscle invasive bladder tumors and undifferentiated bladder cell lines, miR-200 family silencing was associated with DNA hypermethylation (29). Aberrant hypermethylation of miR-34a has been exposed in various hematological malignancies like B-cell chronic lymphocytic leukemia, multiple myeloma and lymphomas, but not in samples from patients with ALL (30). MiR-34 family controls the regulation of Notch1 and Notch 2 protein expressions in glioma cells, pancreatic and prostate tumor cells by interfering the inhibition of self-renewal and differentiation features of Cancer Stem Cells (CSCs) (31). Li et al. for the first time in relation to increased levels of Notch with the miR-34a down-regulation in glioblastoma and medulloblastoma, disclosed that miR-34a directed both Notch-1 and Notch-2 (32). Our study and similar researches on other cancers revealed that hypermethylation of promoter of mir-200b and mir-34a genes on the Notch signal pathway correlate with increased tumor metastases and prognosis of cancer so that demethylation of mir-200b and miR-34 can be eventually used as cancer therapeutic by down-regulating the Notch family members.
In this study patients were treated with drugs like L-asparaginase, vincristine, prednisolone, cytarabine, mercaptopurine, methotrexate, cyclophosphamide and 5-fluorouracil/leucovorin. Hogarth et al. showed that mercaptopurine and cyclophosphamide drugs inhibit DNA methylation (33). But in another study Wang et al. showed that methotrexate and cyclophosphamide drugs cause DNA methylation (34). Also Zhang et al. found that 5-fluorouracil/leucovorin had no effect on methylation in leukemia (35). Moon et al. studied vincristine stimulated demethylation of methylated genes in colorectal tumor cells with 5- azacytidine (AZA) (36). In vitro studies, there are numerous proposals indicating that epigenetic modulation can be significant in the pathogenesis of ALL (37). AZA is a hypomethylating mediator with well-established action in myelodysplastic syndrome and acute myeloid leukemia, but not in ALL (38, 39).
Saiki et al. in 1978 showed the effect of AZA on patients with leukemia and displayed that only 4% of 66 patients with acute lymphoblastic leukemia had recovered perfectly while about 8% responded to treatment. They indicate that the intensity of AZA toxicity, especially in the form of nausea and vomiting, is the main limitation of treatment (40). Hoshino et al. in 2007 demonstrated that Hck tyrosine kinase gene promoter CpG island is aberrantly methylated in leukemia (56.5% in hematopoietic and 80% in non-hematopoietic cell lines) but treatment with 5-aza-2’-deoxycytidine stimulate demethylation of Hck mRNA and protein expression (41). It seems that AZA can be considered as a therapeutic option simultaneously with the administration of anti-leukemia drugs in the treatment of acute lymphoblastic leukemia.
We observed a significant association of mir-200b gene promoter hypermethylation with platelets (P < 0.05) and family history (P < 0.05) in acute lymphoblastic leukemia, also miR-34 gene with cancer state (P < 0.05) and hemoglobin (P < 0.05).
5.1. Conclusions
The methylation of promoter mir200b and mir-34a genes can affect regulation of the key pathways involved in leukemogenesis such as that of Notch pathway. Therefore the use of drugs to reverse epigenetic changes alone or in combination with standard chemotherapy can provide a therapeutic advantage for these patients.