1. Background
Autism spectrum disorder (ASD), a pervasive development disorder, is defined as impairment in social interaction and communication, restricted or repetitive behaviors (1). The onset of ASD usually occurs before the age of three, and the boys are four times more likely to be affected than girls (2). Study showed that the prevalence of ASD was 62/10000 worldwide; statistics indicate the conspicuously increasing prevalence of this disorder over the past 3 decades (3). However, meta-analysis shows that the prevalence of ASD in China is 24.5/10,000 (4). Though the etiology of this disorder remains unclear, a number of studies have proved that ASD is a strong heritable neurodevelopmental disorder (5, 6), and its heritability ranges from 60% to 90% according to families and twins study (7). The concordance of Pairwise ASD was 31% for dizygotic (DZ) and 88% for monozygotic (MZ) twins (8).
Autism spectrum disorder is a complex mental health disorder, and hundreds of genes were found related to it. These related genes are distributed in multiple regions, including 7th, 15th, 17th, and 22nd X chromosomes (9). Also, a number of studies identified that several single-nucleotide polymorphisms (SNP) in different genes had relationship with ASD children. For example, SNPs in tryptophan hydroxylase 2 (TPH2), oxytocin receptor (OXTR), neuroligin1 (NLGN1) and methylenetetrahydrofolate reductase (MTHFR) were all found to have an association with ASD in diverse studies (10-13). In the past three decades, HLA gene complex received increasing attention. Human leukocyte antigen (HLA) gene complex, also known as major histocompatibility complex (MHC), is located on the short arm of 6th chromosome with more than 200 genes in three different regions (class I, class II and class III). Class I region contains gene HLA-A, HLA-B and HLA-C. Class II region contains gene HLA-DRB1, HLA-DQ, and Class III region contains gene HLA-C4A and HLA-C4B. HLA involves a plenty of pathological and physiological responses, including graft immunological rejection, immune response and immune regulation, and some certain autoimmune diseases, such as type I diabetes, ankylosing spondylitis, and systemic lupus erythematosus (SLE). However, several studies demonstrated the association between HLA and ASD. Torres et al. (14) found that HLA-DR is correlated to ASD from its family-based study, and transmission disequilibrium test (TDT) indicated that HLA-A2 allele was significantly associated with ASD. Moreover, other genes in HLA were also reported in some case-control studies. Chien et al. (15) reported of significant association of HLA-DRB1 alleles with ASD in Chinese population. Trajkovski and Spiroski (16) investigated more than 10 alleles of HLA-DRB1 and HLA-C, and observed that 1 allele of each had association with ASD. However, dozens of alleles identified from HLA-1, HLA-C, HLA-DRB1, HLA-DQB1 and haplotypes were all shown having positive relationship with ASD in above articles. Among these, HLA-DRB1 is the most studied one, its allele distributions had been repeatedly reported as a possible gene association with ASD. Additionally, Torres et al. (17) discussed dozens of suspect genes, contained in different regions of HLA gene complex, being also responsible for ASD.
2. Objectives
Since there are no researches about the association of gene HLA-B with ASD in Chinese population, the main purpose of this study was to investigate the association between two SNP of HLA-B and ASD in northeastern Chinese population.
3. Methods
3.1. Ethics Statement
This study passed the approval of the Ethics Committee of School of Public Health, Jilin University (permit no. 2014-05-18), in adherence to the Declaration of Helsinki guidelines. Each subject or their guardians (for children) signed a written informed consent regarding the use of their samples and basic information when they were hospitalized.
3.2. Subjects
Both ASD and healthy children were recruited from Department of Pediatrics, The First Bethune Hospital of Jilin University, during 2011 to 2013. The diagnosis of ASD in these children was confirmed based on the criteria of “diagnostic and statistical manual of mental disorders”, fifth edition (DSM-5). Children with family history of schizophrenia or other mental disorders were excluded from healthy controls. In our case-control study, 84 ASD children and 182 gender and age matched healthy children were enrolled, and 67 family trios were enrolled for family-based study. EpiCalc 2000 software was used to calculate the sample size, detailed information of parameters are reported in our previously published article (18).
3.3. SNP Selection
The SNPs of HAL-B were acquired from the HapMap database (http://www.hapmap.org). We set the parameters for minor allele frequency (MAF) at 0.1, r2 > 0.8, D’ = 1 and two tag SNPs were selected using Haploview version 4.2. Including rs1058026 and rs2770, which are located in the 3’ untranslated region (3’UTR).
3.4. DNA Extraction and Genotyping
Blood samples were preserved in -20°C before following tests. Subjects’ DNA were extracted from peripheral blood using DNA extraction kit (Beijing Kangwei Century Biotech Co, Ltd, China). The concentration and purity of extracted DNA was measured by UV-260 spectrophotometer (Shimadzu, Kyoto, Japan). The threshold of concentration was 15 ng/UL and purity was 1.6 - 1.9. We performed the SNP genotyping using the Sequenom MassARRAY platform (San Diego, CA, USA). The standard polymerase chain reaction (PCR) was performed to amplify DNA. The PCR products were analyzed using the MassARRAY Typer software version 4.0 (Sequenom, USA).
3.5. Statistical Analysis
Case-Control Analysis: Gender and age distribution in cases and controls were compared using chi-square and nonparametric test respectively. Hardy Weinberg equilibrium (HWE) test was performed for each SNP using SNPStats. The differences of allele and genotype frequencies between cases and controls were compared using chi-square test. The best inheritance model was selected according to Akaike information criterion (AIC) and Bayesian information criterion (BIC). The association between haplotypes and ASD was analyzed via SNPstats.
Family-Based Analysis: HWE was detected also before association analysis. Haplotype relative risk test (HRR) and transmission disequilibrium test (TDT) were applied. In HRR, alleles transmitted from parents were defined as transmitted group, and the remaining alleles were defined as non-transmitted group. In TDT, alleles only count if they were transmitted from heterozygous parents, and we assume it is not deviated from TDT if expected frequency of two transmitted alleles meet ratio of 1:1. All statistics were done using R, version 3.4.2 (http://r-project.org). P values < 0.05 are considered as significant.
4. Results
4.1. Case-Control Method
In case-control study, 266 subjects (84 cases, 182 controls) were enrolled. The ratio of male-female was 4.2:1. The median (P25-P75) ages of ASD and healthy children were 4 (3-5). The gender and age distribution were not statistically significant (χ2 = 0.090, P = 0.764 for gender; Z = -0.243, P = 0.808 for age). The genotypic distributions of SNP rs1058026 was not deviated from HWE (P = 0.085), but SNP rs2770 was deviated from HWE (P < 0.001). The results of allelic association analysis is shown in Table 1. Alleles of both SNPs showed no significant differences between cases and controls (rs1058026: χ2 = 3.058, P = 0.080; rs2770: χ2 = 2.965, P = 0.085).
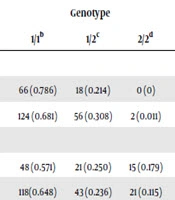
Genotypes of SNP rs1058026 distribution were as follows: T/T 66 (0.786), T/G 18 (0.214), G/G 0 (0) in case group, and T/T 124 (0.681), T/G 56 (0.308), G/G 2 (0.011) in control group. As for SNP rs2770, T/T 48 (0.571), C/T 21 (0.250), C/C 15 (0.179) in case group, and T/T 118 (0.648), T/C 43 (0.236), C/C 21 (0.115) in control group (Table 2). Considering that individuals with GG genotypes were zero, fisher’s exact test was used to analyze the difference of rs1058026 genotypes between two groups, whereas chi-square test was used for rs2770. However, the results of both SNPs were not statistically significant (rs1058026: P = 0.195, rs2770: P = 0.319). Log-additive model was chosen as the fittest model in both SNPS for the smallest values of AIC and BIC, but none of SNPs were found statistically significant under log-additive model.
Haplotype analysis was performed to evaluate the interaction effect of two SNPs, the wild type of each allele was chosen as reference haplotype (TT). No significant associations were found in any haplotypes (Table 3).
| SNP | Group | OR (95% CI) | P | |||
|---|---|---|---|---|---|---|
| rs1058026 | rs2770 | Case | Control | |||
| 1 | T | T | 59.68 | 61.55 | 1.00 | - |
| 2 | T | C | 29.60 | 21.96 | 0.80 (0.55 - 1.16) | 0.24 |
| 3 | G | T | 9.96 | 15.10 | 1.54 (0.81 - 2.94) | 0.19 |
| 4 | G | C | 0.75 | 1.39 | 2.15 (0.23 - 20.01) | 0.50 |
Haplotype Analysis
4.2. Family-Based Method
We excluded two ineligible family trios for rs1058026 and eight for rs2770 due to unmatched genotypes between parents and children. HWE was calculated for two SNPs. Similar to case-control, the genotypic distributions of SNP rs1058026 was not deviated from HWE (P = 0.773), and SNP rs2770 was deviated from HWE (P < 0.001).
Table 4 shows that the T allele frequencies of SNP rs1058026 in transmitted group were significantly higher than those in non-transmitted group [0.9% (T) vs 0.792% (NT); χ2 = 5.790, P = 0.016)]. However, significant differences of alleles were not observed in SNP rs2770 (χ2 = 0.091, P = 0.762).
| SNP | Allele | T | NT | χ2 | P |
|---|---|---|---|---|---|
| rs1058026 | T | 117 (0.9) | 103 (0.792) | 5.790 | 0.016 |
| G | 13 (0.1) | 27 (0.208) | |||
| rs2770 | T | 90 (0.763) | 88 (0.746) | 0.091 | 0.762 |
| C | 28 (0.237) | 30 (0.254) |
HRR Analysisa
In TDT analysis, the frequencies of allele T was significantly higher than G allele in SNP rs1058026 [25(T) vs. 11(G); χ2 = 5.444, P = 0.020]. Whereas no significant difference was found in SNP rs2770 (χ2 = 0.024, P = 0.876; Table 5).
| SNP | Allele | Transmitted | χ2 | P |
|---|---|---|---|---|
| rs1058026 | T | 25 | 5.444 | 0.020 |
| G | 11 | |||
| rs2770 | T | 20 | 0.024 | 0.876 |
| C | 21 |
TDT Analysis
5. Discussion
The present study attempts to determine the association of HLA-B polymorphisms with ASD in northeast Chinese population by using case-control and family-based method. In our case-control study, the ratio of male/female was 4.2:1 which was consistent with prevalence of ASD. Then, we calculated the allele frequency, genotype distributions and analyzed its relationships with two SNPs. Haplotype analysis was also performed to evaluate SNPs interaction. None of above analyses came out with positive results. In contrast, we found that rs1058026 was significantly associated with ASD in both family-based methods (HRR and TDT), and allele T was preferentially transmitted to ASD children.
HWE is a state in which allele and genotype frequencies in a population remains constant from generation to generation in the absence of other evolutionary influences, and HWE test has long been recommend as the first step to be carried out in genetic analysis. Recently, some researchers believe that a population will never be exactly in HWE, it should not be used to identify genotyping errors and it is only valid with a very large sample size (19-21). Although the SNP rs2770 is derived from HWE in our study, we do not rule it out.
HLA proteins transmit antigens to two specific immune cells, namely T cells and B cells. T cells eliminate the antigens by the cytotoxic effect, whereas B cells produce antibodies and cytokines. These immune responses are also known as cellular and humoral immunity. However, the phenomenon of altered immune function and immune dysregulation in individuals with ASD was observed. The elevated level of inflammatory cytokines (TNF, IL) (22, 23), along with abnormal expression of chemokines (MCP-1, MIP-1α, RANTES) in plasma (24) as well as altered levels of multiple immune cells (T cells, B cells, NK cells) were found in ASD (25, 26). It is worth noting that such immune responses will eventually account for inflammations, especially in brain. The above-mentioned findings suggest that immune dysfunction plays a pivotal role in pathogenesis of ASD. Hence, it is believed that understanding these alterations will help us to identify ASD in an early stage and initiate a better treatment.
Several studies have evaluated the correlation of allele frequencies and genotype distributions of HLA-B gene with ASD. Apichaya found six alleles of HLA-B correlated with ASD in Thai population, including HLA-B∗13:02, HLA-B∗38:02, HLA-B∗44:03, HLA-B∗56:01, HLA-B∗18:02, HLA-B∗46:12. He suggested that HLA-B∗18:02, HLA-B∗46:12 might play a protective role in ASD (27). Interesting enough, same alleles were selected in another study, but none of these alleles showed consistent results (16). Al-Hakbany et al. (28) could only find HLA-B∗07 significantly associated with ASD in his case-control study, the extended haplotype HLA-A∗02-B∗07 was significantly higher in ASD compared to controls. The reason that causes these conflicting results could be complex. First, sample sizes were considerably small due to low prevalence. Moreover, it is tricky to diagnose ASD and distinguish from other neurodevelopmental disorders. Second, the diverse ethnics and regions of subjects could contribute to differential allele distribution. In another study, the haplotype B44-SC30-DR4, TNF-238(G). TNF-308 (G), MIB*332, HLA-B*38, and HLA-Cw*12 were found associated with ASD in haplotype analysis (29, 30). This indicates that gene HLA-B might take a part in developing ASD by combining with other genes.
The major limitation of this study was the difficulty of subject collection which accounts for a small sample size. However, since the family-based study has a better statistic power and avoids some certain confounding factors such as false match issue occurred in case-control study and spurious association caused by population stratification, according to the results from HRR and TDT, we still have reasons to believe the association found in family-based study is significant.
In summary, SNP rs1058026 was never studied before to our best knowledge. Accordingly, the results obtained from our study will provide fundamental evidences for possible association between gene HLA-B and children with ASD. However, rs1058026 of HLA-B was found to be associated with ASD in our study.